GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
TRAF2 and NCK interacting kinase
Target id: 2244
Nomenclature: TRAF2 and NCK interacting kinase
Abbreviated Name: TNIK
Family: MSN subfamily
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 1360 | 3q26.2-q26.31 | TNIK | TRAF2 and NCK interacting kinase | |
| Mouse | - | 1323 | 3 A3 | Tnik | TRAF2 and NCK interacting kinase | |
| Rat | - | 1354 | 2 q24 | Tnik | TRAF2 and NCK interacting kinase | |
Database Links  |
|
| Alphafold | Q9UKE5 (Hs), P83510 (Mm) |
| BRENDA | 2.7.11.1 |
| ChEMBL Target | CHEMBL4527 (Hs) |
| Ensembl Gene | ENSG00000154310 (Hs), ENSMUSG00000027692 (Mm), ENSRNOG00000012422 (Rn) |
| Entrez Gene | 23043 (Hs), 665113 (Mm), 294917 (Rn) |
| Human Protein Atlas | ENSG00000154310 (Hs) |
| KEGG Enzyme | 2.7.11.1 |
| KEGG Gene | hsa:23043 (Hs), mmu:665113 (Mm), rno:294917 (Rn) |
| OMIM | 610005 (Hs) |
| Pharos | Q9UKE5 (Hs) |
| RefSeq Nucleotide | NM_001161560 (Hs), NM_001163007 (Mm), NM_001106422 (Rn) |
| RefSeq Protein | NP_001155032 (Hs), NP_001156479 (Mm), NP_001099892 (Rn) |
| UniProtKB | Q9UKE5 (Hs), P83510 (Mm) |
| Wikipedia | TNIK (Hs) |
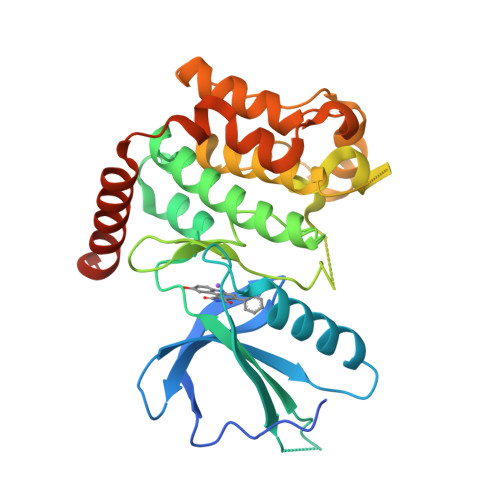
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
Enzyme Reaction  |
||||
|
||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
DiscoveRx KINOMEscan® screen  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen of 72 inhibitors against 456 human kinases. Quantitative data were derived using DiscoveRx KINOMEscan® platform. http://www.discoverx.com/services/drug-discovery-development-services/kinase-profiling/kinomescan Reference: 2,7 |
 
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: TNIK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
1. Ammirati M, Bagley SW, Bhattacharya SK, Buckbinder L, Carlo AA, Conrad R, Cortes C, Dow RL, Dowling MS, El-Kattan A et al.. (2015) Discovery of an in Vivo Tool to Establish Proof-of-Concept for MAP4K4-Based Antidiabetic Treatment. ACS Med Chem Lett, 6 (11): 1128-33. [PMID:26617966]
2. Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1046-51. [PMID:22037378]
3. Ho KK, Parnell KM, Yuan Y, Xu Y, Kultgen SG, Hamblin S, Hendrickson TF, Luo B, Foulks JM, McCullar MV et al.. (2013) Discovery of 4-phenyl-2-phenylaminopyridine based TNIK inhibitors. Bioorg Med Chem Lett, 23 (2): 569-73. [PMID:23232060]
4. Li Y, Zhang L, Yang R, Qiao Z, Wu M, Huang C, Tian C, Luo X, Yang W, Zhang Y et al.. (2022) Discovery of 3,4-Dihydrobenzo[f][1,4]oxazepin-5(2H)-one Derivatives as a New Class of Selective TNIK Inhibitors and Evaluation of Their Anti-Colorectal Cancer Effects. J Med Chem, 65 (3): 1786-1807. [PMID:34985886]
5. Ren F, Aliper A, Chen J, Zhao H, Rao S, Kuppe C, Ozerov IV, Zhang M, Witte K, Kruse C et al.. (2025) A small-molecule TNIK inhibitor targets fibrosis in preclinical and clinical models. Nat Biotechnol, 43 (1): 63-75. [PMID:38459338]
6. Shuai W, Bu F, Zhu Y, Wu Y, Xiao H, Pan X, Zhang J, Sun Q, Wang G, Ouyang L. (2023) Discovery of Novel Indazole Chemotypes as Isoform-Selective JNK3 Inhibitors for the Treatment of Parkinson's Disease. J Med Chem, 66 (2): 1273-1300. [PMID:36649216]
7. Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP et al.. (2010) Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem Biol, 17 (11): 1241-9. [PMID:21095574]
How to cite this page
MSN subfamily: TRAF2 and NCK interacting kinase. Last modified on 03/06/2025. Accessed on 11/02/2026. IUPHAR/BPS Guide to PHARMACOLOGY, https://www.guidetomalariapharmacology.org/GRAC/ObjectDisplayForward?objectId=2244.