GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
mitogen-activated protein kinase kinase 6
Target id: 2067
Nomenclature: mitogen-activated protein kinase kinase 6
Abbreviated Name: MKK6
Family: STE7 family
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 334 | 17q24.3 | MAP2K6 | mitogen-activated protein kinase kinase 6 | |
| Mouse | - | 334 | 11 E1-E2 | Map2k6 | mitogen-activated protein kinase kinase 6 | |
| Rat | - | 334 | 10q32.1 | Map2k6 | mitogen-activated protein kinase kinase 6 | |
Database Links  |
|
| Alphafold | P52564 (Hs), P70236 (Mm), Q925D6 (Rn) |
| BRENDA | 2.7.12.2 |
| ChEMBL Target | CHEMBL2171 (Hs) |
| Ensembl Gene | ENSG00000108984 (Hs), ENSMUSG00000020623 (Mm), ENSRNOG00000004437 (Rn) |
| Entrez Gene | 5608 (Hs), 26399 (Mm), 114495 (Rn) |
| Human Protein Atlas | ENSG00000108984 (Hs) |
| KEGG Enzyme | 2.7.12.2 |
| KEGG Gene | hsa:5608 (Hs), mmu:26399 (Mm), rno:114495 (Rn) |
| OMIM | 601254 (Hs) |
| Pharos | P52564 (Hs) |
| RefSeq Nucleotide | NM_002758 (Hs), NM_011943 (Mm), NM_053703 (Rn) |
| RefSeq Protein | NP_002749 (Hs), NP_036073 (Mm), NP_446155 (Rn) |
| SynPHARM | 81546 (in complex with staurosporine) |
| UniProtKB | P52564 (Hs), P70236 (Mm), Q925D6 (Rn) |
| Wikipedia | MAP2K6 (Hs) |
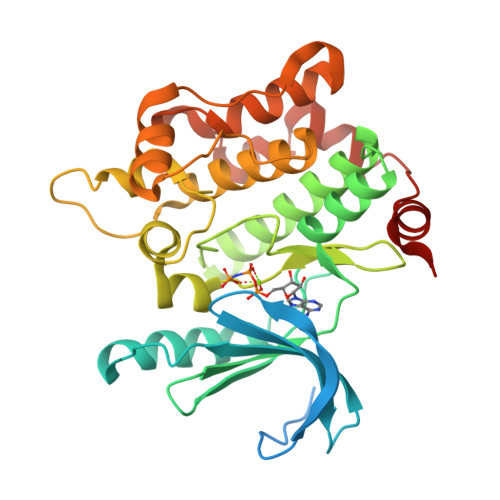
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
Enzyme Reaction  |
||||
|
||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
DiscoveRx KINOMEscan® screen  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen of 72 inhibitors against 456 human kinases. Quantitative data were derived using DiscoveRx KINOMEscan® platform. http://www.discoverx.com/services/drug-discovery-development-services/kinase-profiling/kinomescan Reference: 2,6 |
 
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: MEK6 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
EMD Millipore KinaseProfilerTM screen/Reaction Biology Kinase HotspotSM screen  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen profiling 158 kinase inhibitors (Calbiochem Protein Kinase Inhibitor Library I and II, catalogue numbers 539744 and 539745) for their inhibitory activity at 1µM and 10µM against 234 human recombinant kinases using the EMD Millipore KinaseProfilerTM service. A screen profiling the inhibitory activity of 178 commercially available kinase inhibitors at 0.5µM against a panel of 300 recombinant protein kinases using the Reaction Biology Corporation Kinase HotspotSM platform. http://www.millipore.com/techpublications/tech1/pf3036 http://www.reactionbiology.com/webapps/main/pages/kinase.aspx Reference: 1,3 |
 
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: MKK6/MKK6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
1. Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1039-45. [PMID:22037377]
2. Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1046-51. [PMID:22037378]
3. Gao Y, Davies SP, Augustin M, Woodward A, Patel UA, Kovelman R, Harvey KJ. (2013) A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochem J, 451 (2): 313-28. [PMID:23398362]
4. Matsumoto T, Kinoshita T, Matsuzaka H, Nakai R, Kirii Y, Yokota K, Tada T. (2012) Crystal structure of non-phosphorylated MAP2K6 in a putative auto-inhibition state. J Biochem, 151 (5): 541-9. [PMID:22383536]
5. Pandey A, Volkots DL, Seroogy JM, Rose JW, Yu JC, Lambing JL, Hutchaleelaha A, Hollenbach SJ, Abe K, Giese NA et al.. (2002) Identification of orally active, potent, and selective 4-piperazinylquinazolines as antagonists of the platelet-derived growth factor receptor tyrosine kinase family. J Med Chem, 45 (17): 3772-93. [PMID:12166950]
6. Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP et al.. (2010) Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem Biol, 17 (11): 1241-9. [PMID:21095574]
How to cite this page
STE7 family: mitogen-activated protein kinase kinase 6. Last modified on 12/02/2015. Accessed on 30/01/2026. IUPHAR/BPS Guide to PHARMACOLOGY, https://www.guidetomalariapharmacology.org/GRAC/ObjectDisplayForward?objectId=2067.