GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
MER proto-oncogene, tyrosine kinase
Target id: 1837
Nomenclature: MER proto-oncogene, tyrosine kinase
Abbreviated Name: Mer
Family: Type XI RTKs: TAM (TYRO3-, AXL- and MER-TK) receptor family
Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Enzyme Reaction
- Natural/Endogenous Ligands
- Inhibitors
- Large-scale Ligand Screening Data
- Immunopharmacology Comments
- Immuno Cell Type Associations
- Clinically-Relevant Mutations and Pathophysiology
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 1 | 999 | 2q13 | MERTK | MER proto-oncogene, tyrosine kinase | |
| Mouse | 1 | 994 | 2 F1 | Mertk | MER proto-oncogene tyrosine kinase | |
| Rat | 1 | 994 | 3q36 | Mertk | MER proto-oncogene, tyrosine kinase | |
Previous and Unofficial Names  |
| RP38 | Rdy | retinal dystrophy | tyrosine-protein kinase Mer | Eyk | Nyk | Tyro 12 | c-mer proto-oncogene tyrosine kinase | MER proto-oncogene |
Database Links  |
|
| Alphafold | Q12866 (Hs), Q60805 (Mm), P57097 (Rn) |
| BRENDA | 2.7.10.1 |
| CATH/Gene3D | 2.60.40.10 |
| ChEMBL Target | CHEMBL5331 (Hs), CHEMBL4879469 (Mm) |
| Ensembl Gene | ENSG00000153208 (Hs), ENSMUSG00000014361 (Mm), ENSRNOG00000017319 (Rn) |
| Entrez Gene | 10461 (Hs), 17289 (Mm), 65037 (Rn) |
| Human Protein Atlas | ENSG00000153208 (Hs) |
| KEGG Enzyme | 2.7.10.1 |
| KEGG Gene | hsa:10461 (Hs), mmu:17289 (Mm), rno:65037 (Rn) |
| OMIM | 604705 (Hs) |
| Orphanet | ORPHA123198 (Hs) |
| Pharos | Q12866 (Hs) |
| RefSeq Nucleotide | NM_006343 (Hs), NM_008587 (Mm), NM_022943 (Rn) |
| RefSeq Protein | NP_006334 (Hs), NP_032613 (Mm), NP_075232 (Rn) |
| UniProtKB | Q12866 (Hs), Q60805 (Mm), P57097 (Rn) |
| Wikipedia | MERTK (Hs) |
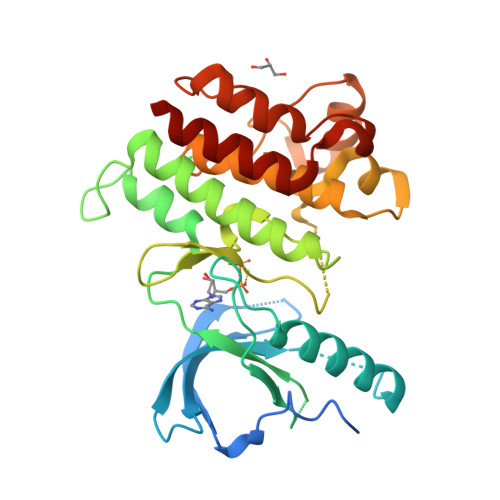
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
Enzyme Reaction  |
||||
|
||||
Natural/Endogenous Ligands  |
| growth arrest specific protein 6 {Sp: Human} |
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
DiscoveRx KINOMEscan® screen  |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen of 72 inhibitors against 456 human kinases. Quantitative data were derived using DiscoveRx KINOMEscan® platform. http://www.discoverx.com/services/drug-discovery-development-services/kinase-profiling/kinomescan Reference: 5,23 |
 
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: MERTK | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
EMD Millipore KinaseProfilerTM screen/Reaction Biology Kinase HotspotSM screen  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
A screen profiling 158 kinase inhibitors (Calbiochem Protein Kinase Inhibitor Library I and II, catalogue numbers 539744 and 539745) for their inhibitory activity at 1µM and 10µM against 234 human recombinant kinases using the EMD Millipore KinaseProfilerTM service. A screen profiling the inhibitory activity of 178 commercially available kinase inhibitors at 0.5µM against a panel of 300 recombinant protein kinases using the Reaction Biology Corporation Kinase HotspotSM platform. http://www.millipore.com/techpublications/tech1/pf3036 http://www.reactionbiology.com/webapps/main/pages/kinase.aspx Reference: 1,6 |
 
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Target used in screen: Mer/c-MER | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displaying the top 10 most potent ligands View all ligands in screen » | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Immunopharmacology Comments |
| Mer plays a critical role in regulating self-tolerance mediated between apoptotic cells, dendritic cells, and T cells [4,22]. |
| Cell Type Associations | ||||||||
|
||||||||
|
||||||||
|
||||||||
|
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||||||
|
||||||||||||||
References
1. Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. (2011) Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1039-45. [PMID:22037377]
2. Bae SH, Kim JH, Park TH, Lee K, Lee BI, Jang H. (2022) BMS794833 inhibits macrophage efferocytosis by directly binding to MERTK and inhibiting its activity. Exp Mol Med, 54 (9): 1450-1460. [PMID:36056187]
3. Bannen LC, Bui M, Jiang F, Tso K, Wang Y, Xu W. (2019) Compounds for the treatment of kinase-dependent disorders. Patent number: WO2019148044A1. Assignee: Exelixis, Inc.. Priority date: 26/01/2018. Publication date: 01/08/2019.
4. Behrens EM, Gadue P, Gong SY, Garrett S, Stein PL, Cohen PL. (2003) The mer receptor tyrosine kinase: expression and function suggest a role in innate immunity. Eur J Immunol, 33 (8): 2160-7. [PMID:12884290]
5. Davis MI, Hunt JP, Herrgard S, Ciceri P, Wodicka LM, Pallares G, Hocker M, Treiber DK, Zarrinkar PP. (2011) Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol, 29 (11): 1046-51. [PMID:22037378]
6. Gao Y, Davies SP, Augustin M, Woodward A, Patel UA, Kovelman R, Harvey KJ. (2013) A broad activity screen in support of a chemogenomic map for kinase signalling research and drug discovery. Biochem J, 451 (2): 313-28. [PMID:23398362]
7. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S et al.. (2012) Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol, 13 (11): 1118-28. [PMID:23023392]
8. Huang X, Finerty P, Walker JR, Butler-Cole C, Vedadi M, Schapira M, Parker SA, Turk BE, Thompson DA, Dhe-Paganon S. (2009) Structural insights into the inhibited states of the Mer receptor tyrosine kinase. J Struct Biol, 165 (2): 88-96. [PMID:19028587]
9. Kong D, Tian Q, Chen Z, Zheng H, Stashko MA, Yan D, Earp HS, Frye SV, DeRyckere D, Kireev D et al.. (2024) Discovery of Novel Macrocyclic MERTK/AXL Dual Inhibitors. J Med Chem, 67 (7): 5866-5882. [PMID:38556760]
10. Lee HK, Kim HW, Lee IY, Lee J, Lee J, Jung DS, Lee SY, Park SH, Hwang H, Choi JS et al.. (2014) G-749, a novel FLT3 kinase inhibitor, can overcome drug resistance for the treatment of acute myeloid leukemia. Blood, 123 (14): 2209-19. [PMID:24532805]
11. Lee LY, Hernandez D, Rajkhowa T, Smith SC, Raman JR, Nguyen B, Small D, Levis M. (2017) Preclinical studies of gilteritinib, a next-generation FLT3 inhibitor. Blood, 129 (2): 257-260. [PMID:27908881]
12. Lewis RT, Bode CM, Choquette DM, Potashman M, Romero K, Stellwagen JC, Teffera Y, Moore E, Whittington DA, Chen H et al.. (2012) The discovery and optimization of a novel class of potent, selective, and orally bioavailable anaplastic lymphoma kinase (ALK) inhibitors with potential utility for the treatment of cancer. J Med Chem, 55 (14): 6523-40. [PMID:22734674]
13. Li Y, Xiong Y, Zhang G, Zhang L, Yang W, Yang J, Huang L, Qiao Z, Miao Z, Lin G et al.. (2018) Identification of 5-(2,3-Dihydro-1 H-indol-5-yl)-7 H-pyrrolo[2,3- d]pyrimidin-4-amine Derivatives as a New Class of Receptor-Interacting Protein Kinase 1 (RIPK1) Inhibitors, Which Showed Potent Activity in a Tumor Metastasis Model. J Med Chem, 61 (24): 11398-11414. [PMID:30480444]
14. Nam K, Kim J, Park D, Jeon Y, Yang YI, Kang HK. (2021) Quinoline derivatives as inhibitors of axl/mer rtk and csf1r. Patent number: US20210163448A1. Assignee: QURIENT CO Ltd. Priority date: 31/05/2019. Publication date: 03/06/2021.
15. Pan BS, Chan GK, Chenard M, Chi A, Davis LJ, Deshmukh SV, Gibbs JB, Gil S, Hang G, Hatch H et al.. (2010) MK-2461, a novel multitargeted kinase inhibitor, preferentially inhibits the activated c-Met receptor. Cancer Res, 70 (4): 1524-33. [PMID:20145145]
16. Paolino M, Choidas A, Wallner S, Pranjic B, Uribesalgo I, Loeser S, Jamieson AM, Langdon WY, Ikeda F, Fededa JP et al.. (2014) The E3 ligase Cbl-b and TAM receptors regulate cancer metastasis via natural killer cells. Nature, 507 (7493): 508-12. [PMID:24553136]
17. Patwardhan PP, Ivy KS, Musi E, de Stanchina E, Schwartz GK. (2016) Significant blockade of multiple receptor tyrosine kinases by MGCD516 (Sitravatinib), a novel small molecule inhibitor, shows potent anti-tumor activity in preclinical models of sarcoma. Oncotarget, 7 (4): 4093-109. [PMID:26675259]
18. Peng YH, Li MC, Yen WC, Yeh TK, Hsueh CC, Kuo FM, Lai YL, Chang L, Lee LC, Chen PY et al.. (2025) Structure-Based Design of Potent and Selective MerTK Inhibitors by Modulating the Conformation of αC Helix. J Med Chem, 68 (11): 10877-10896. [PMID:40391976]
19. Rothlin CV, Ghosh S, Zuniga EI, Oldstone MB, Lemke G. (2007) TAM receptors are pleiotropic inhibitors of the innate immune response. Cell, 131 (6): 1124-36. [PMID:18083102]
20. Schroeder GM, An Y, Cai ZW, Chen XT, Clark C, Cornelius LA, Dai J, Gullo-Brown J, Gupta A, Henley B et al.. (2009) Discovery of N-(4-(2-amino-3-chloropyridin-4-yloxy)-3-fluorophenyl)-4-ethoxy-1-(4-fluorophenyl)-2-oxo-1,2-dihydropyridine-3-carboxamide (BMS-777607), a selective and orally efficacious inhibitor of the Met kinase superfamily. J Med Chem, 52 (5): 1251-4. [PMID:19260711]
21. Shao WH, Zhen Y, Eisenberg RA, Cohen PL. (2009) The Mer receptor tyrosine kinase is expressed on discrete macrophage subpopulations and mainly uses Gas6 as its ligand for uptake of apoptotic cells. Clin Immunol, 133 (1): 138-44. [PMID:19631584]
22. Wallet MA, Sen P, Flores RR, Wang Y, Yi Z, Huang Y, Mathews CE, Earp HS, Matsushima G, Wang B et al.. (2008) MerTK is required for apoptotic cell-induced T cell tolerance. J Exp Med, 205 (1): 219-32. [PMID:18195070]
23. Wodicka LM, Ciceri P, Davis MI, Hunt JP, Floyd M, Salerno S, Hua XH, Ford JM, Armstrong RC, Zarrinkar PP et al.. (2010) Activation state-dependent binding of small molecule kinase inhibitors: structural insights from biochemistry. Chem Biol, 17 (11): 1241-9. [PMID:21095574]
24. Yan SB, Peek VL, Ajamie R, Buchanan SG, Graff JR, Heidler SA, Hui YH, Huss KL, Konicek BW, Manro JR et al.. (2013) LY2801653 is an orally bioavailable multi-kinase inhibitor with potent activity against MET, MST1R, and other oncoproteins, and displays anti-tumor activities in mouse xenograft models. Invest New Drugs, 31 (4): 833-44. [PMID:23275061]
25. Yu Y, Jang M, Miyashiro J, Clark RF, Zhu GD, Gong J, Dai Y, Frey RR, Penning TD, Kim H et al.. (2024) Discovery of A-910, a Highly Potent and Orally Bioavailable Dual MerTK/Axl-Selective Tyrosine Kinase Inhibitor. J Med Chem, 67 (19): 17000-17032. [PMID:39283694]
26. Zhang Z. (2015) Aminopyridine derivatives as tam family kinase inhibitors. Patent number: WO2015081257A2. Assignee: Signalchem Lifesciences Corporation. Priority date: 26/11/2014. Publication date: 04/06/2015.
27. Zhao J, Zhang D, Zhang W, Stashko MA, DeRyckere D, Vasileiadi E, Parker RE, Hunter D, Liu Q, Zhang Y et al.. (2018) Highly Selective MERTK Inhibitors Achieved by a Single Methyl Group. J Med Chem, 61 (22): 10242-10254. [PMID:30347155]