GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
Synonyms: RAS Inhibitor A122 | RMC-6236 | RMC6236
Compound class:
Synthetic organic
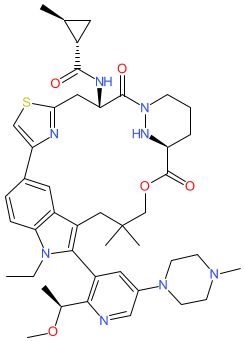
Comment: RMC-6236 is a macrocyclic, noncovalent (reversible) pan-RAS inhibitor [1]. It was developed using structure-guided design from the bacterial polyketide Sanglifehrin A. RMC-6236 forms a binary complex with the ubiquitously-expressed chaperone protein cyclophilin A. The binary complex has high affinity for RAS proteins, and interactions lead to the formation of an inhibitory tri-complex. RMC-6236 has activity against active state (GTP-bound) mutant and wild-type RAS proteins. This mode of action is described as RAS(ON) inhibition, and is intended to target tumours with oncogenic RAS mutations beyond the common driver mutation KRASG12C [1]. The chemical structure of RMC-6236 is identical to that for the INN daraxonrasib (proposed list 132, Feb. 2025).
Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖ |
|
|||||||||||||||||||||||||||||||||||