GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
regulator of G-protein signaling 4
Target id: 2811
Nomenclature: regulator of G-protein signaling 4
Abbreviated Name: RGS4
Family: R4 family
Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Associated Proteins
- Inhibitors
- Tissue Distribution
- Functional Assays
- Physiological Functions
- Physiological Consequences of Altering Gene Expression
- Xenobiotics Influencing Gene Expression
- Clinically-Relevant Mutations and Pathophysiology
- Biologically Significant Variants
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 205 | 1q23.3 | RGS4 | regulator of G protein signaling 4 | |
| Mouse | - | 205 | 1 76.84 cM | Rgs4 | regulator of G-protein signaling 4 | |
| Rat | - | 205 | 13q24 | Rgs4 | regulator of G-protein signaling 4 | |
Previous and Unofficial Names  |
| ESTM48 | ESTM50 |
Database Links  |
|
| Alphafold | P49798 (Hs), O08899 (Mm), P49799 (Rn) |
| CATH/Gene3D | 1.10.196.10 |
| ChEMBL Target | CHEMBL1795091 (Hs) |
| Ensembl Gene | ENSG00000117152 (Hs), ENSMUSG00000038530 (Mm), ENSRNOG00000002773 (Rn) |
| Entrez Gene | 5999 (Hs), 19736 (Mm), 29480 (Rn) |
| Human Protein Atlas | ENSG00000117152 (Hs) |
| KEGG Gene | hsa:5999 (Hs), mmu:19736 (Mm), rno:29480 (Rn) |
| OMIM | 602516 (Hs) |
| Pharos | P49798 (Hs) |
| RefSeq Nucleotide | NM_001102445 (Hs), NM_009062 (Mm), NM_017214 (Rn) |
| RefSeq Protein | NP_001106851 (Hs), NP_001095915 (Hs), NP_033088 (Mm), NP_058910 (Rn) |
| UniProtKB | P49798 (Hs), O08899 (Mm), P49799 (Rn) |
| Wikipedia | RGS4 (Hs) |
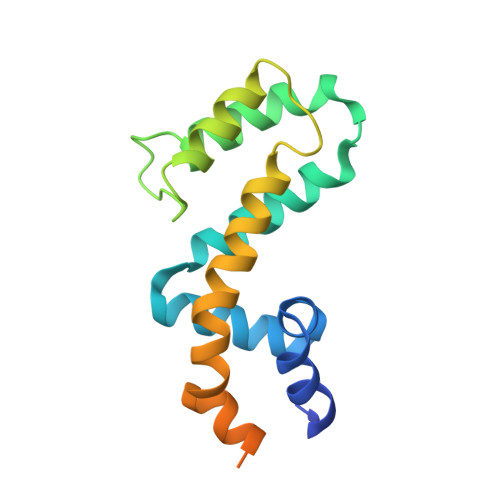
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
Associated Proteins  |
||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||||||||||||||||||||||||||
| Associated Protein Comments | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Affinity for Gαq is lower than that for Gαi. | ||||||||||||||||||||||||||||||||||||||||||||||||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Functional Assays 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Xenobiotics Influencing Gene Expression 
|
||||||||||
|
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||||||
|
||||||||||||||
|
||||||||||||||
Biologically Significant Variants 
|
||||||||||||
|
||||||||||||
|
||||||||||||
|
References
1. Avrampou K, Pryce KD, Ramakrishnan A, Sakloth F, Gaspari S, Serafini RA, Mitsi V, Polizu C, Swartz C, Ligas B et al.. (2019) RGS4 Maintains Chronic Pain Symptoms in Rodent Models. J Neurosci, 39 (42): 8291-8304. [PMID:31308097]
2. Berman DM, Kozasa T, Gilman AG. (1996) The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J Biol Chem, 271 (44): 27209-12. [PMID:8910288]
3. Berman DM, Wilkie TM, Gilman AG. (1996) GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell, 86 (3): 445-52. [PMID:8756726]
4. Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. (2010) Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol, 78 (3): 524-33. [PMID:20571077]
5. Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. (2011) A nanomolar-potency small molecule inhibitor of regulator of G-protein signaling proteins. Biochemistry, 50 (15): 3181-92. [PMID:21329361]
6. Bodenstein J, Sunahara RK, Neubig RR. (2007) N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol, 71 (4): 1040-50. [PMID:17220356]
7. Cervantes-Villagrana RD, Adame-García SR, García-Jiménez I, Color-Aparicio VM, Beltrán-Navarro YM, König GM, Kostenis E, Reyes-Cruz G, Gutkind JS, Vázquez-Prado J. (2019) Gβγ signaling to the chemotactic effector P-REX1 and mammalian cell migration is directly regulated by Gαq and Gα13 proteins. J Biol Chem, 294 (2): 531-546. [PMID:30446620]
8. Chen X, Dunham C, Kendler S, Wang X, O'Neill FA, Walsh D, Kendler KS. (2004) Regulator of G-protein signaling 4 (RGS4) gene is associated with schizophrenia in Irish high density families. Am J Med Genet B Neuropsychiatr Genet, 129B (1): 23-6. [PMID:15274033]
9. Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, B K T, Ferrell RE, Middleton FA et al.. (2002) Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet, 11 (12): 1373-80. [PMID:12023979]
10. Chuang HH, Yu M, Jan YN, Jan LY. (1998) Evidence that the nucleotide exchange and hydrolysis cycle of G proteins causes acute desensitization of G-protein gated inward rectifier K+ channels. Proc Natl Acad Sci USA, 95 (20): 11727-32. [PMID:9751733]
11. Cifelli C, Rose RA, Zhang H, Voigtlaender-Bolz J, Bolz SS, Backx PH, Heximer SP. (2008) RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ Res, 103 (5): 527-35. [PMID:18658048]
12. Ding L, Hegde AN. (2009) Expression of RGS4 splice variants in dorsolateral prefrontal cortex of schizophrenic and bipolar disorder patients. Biol Psychiatry, 65 (6): 541-5. [PMID:19041089]
13. Dowal L, Elliott J, Popov S, Wilkie TM, Scarlata S. (2001) Determination of the contact energies between a regulator of G protein signaling and G protein subunits and phospholipase C beta 1. Biochemistry, 40 (2): 414-21. [PMID:11148035]
14. Ehses J, Fernández-Moya SM, Schröger L, Kiebler MA. (2021) Synergistic regulation of Rgs4 mRNA by HuR and miR-26/RISC in neurons. RNA Biol, 18 (7): 988-998. [PMID:32779957]
15. Garzón J, Rodríguez-Muñoz M, de la Torre-Madrid E, Sánchez-Blázquez P. (2005) Effector antagonism by the regulators of G protein signalling (RGS) proteins causes desensitization of mu-opioid receptors in the CNS. Psychopharmacology (Berl.), 180 (1): 1-11. [PMID:15830230]
16. Georgoussi Z, Leontiadis L, Mazarakou G, Merkouris M, Hyde K, Hamm H. (2006) Selective interactions between G protein subunits and RGS4 with the C-terminal domains of the mu- and delta-opioid receptors regulate opioid receptor signaling. Cell Signal, 18 (6): 771-82. [PMID:16120478]
17. Gold SJ, Ni YG, Dohlman HG, Nestler EJ. (1997) Regulators of G-protein signaling (RGS) proteins: region-specific expression of nine subtypes in rat brain. J Neurosci, 17 (20): 8024-37. [PMID:9315921]
18. Grillet N, Pattyn A, Contet C, Kieffer BL, Goridis C, Brunet JF. (2005) Generation and characterization of Rgs4 mutant mice. Mol Cell Biol, 25 (10): 4221-8. [PMID:15870291]
19. Guda MR, Velpula KK, Asuthkar S, Cain CP, Tsung AJ. (2020) Targeting RGS4 Ablates Glioblastoma Proliferation. Int J Mol Sci, 21 (9). [PMID:32392739]
20. He Z, Yu L, Luo S, Li Q, Huang S, An Y. (2019) RGS4 Regulates Proliferation And Apoptosis Of NSCLC Cells Via microRNA-16 And Brain-Derived Neurotrophic Factor. Onco Targets Ther, 12: 8701-8714. [PMID:31695428]
21. Heximer SP, Watson N, Linder ME, Blumer KJ, Hepler JR. (1997) RGS2/G0S8 is a selective inhibitor of Gqalpha function. Proc Natl Acad Sci USA, 94 (26): 14389-93. [PMID:9405622]
22. Huang MW, Lin YJ, Chang CW, Lei FJ, Ho EP, Liu RS, Shyu WC, Hsieh CH. (2018) RGS4 deficit in prefrontal cortex contributes to the behaviors related to schizophrenia via system xc--mediated glutamatergic dysfunction in mice. Theranostics, 8 (17): 4781-4794. [PMID:30279737]
23. Jaén C, Doupnik CA. (2006) RGS3 and RGS4 differentially associate with G protein-coupled receptor-Kir3 channel signaling complexes revealing two modes of RGS modulation. Precoupling and collision coupling. J Biol Chem, 281 (45): 34549-60. [PMID:16973624]
24. Jin Y, Zhong H, Omnaas JR, Neubig RR, Mosberg HI. (2004) Structure-based design, synthesis, and pharmacologic evaluation of peptide RGS4 inhibitors. J Pept Res, 63 (2): 141-6. [PMID:15009535]
25. Karoussiotis C, Marti-Solano M, Stepniewski TM, Symeonof A, Selent J, Georgoussi Z. (2020) A highly conserved δ-opioid receptor region determines RGS4 interaction. FEBS J, 287 (4): 736-748. [PMID:31386272]
26. Kim J, Lee S, Kang S, Jeon TI, Kang MJ, Lee TH, Kim YS, Kim KS, Im HI, Moon C. (2018) Regulator of G-Protein Signaling 4 (RGS4) Controls Morphine Reward by Glutamate Receptor Activation in the Nucleus Accumbens of Mouse Brain. Mol Cells, 41 (5): 454-464. [PMID:29754475]
27. Kim Y, Ghil S. (2020) Regulators of G-protein signaling, RGS2 and RGS4, inhibit protease-activated receptor 4-mediated signaling by forming a complex with the receptor and Gα in live cells. Cell Commun Signal, 18 (1): 86. [PMID:32517689]
28. Kimple AJ, Willard FS, Giguère PM, Johnston CA, Mocanu V, Siderovski DP. (2007) The RGS protein inhibitor CCG-4986 is a covalent modifier of the RGS4 Galpha-interaction face. Biochim Biophys Acta, 1774 (9): 1213-20. [PMID:17660054]
29. Ko WK, Martin-Negrier ML, Bezard E, Crossman AR, Ravenscroft P. (2014) RGS4 is involved in the generation of abnormal involuntary movements in the unilateral 6-OHDA-lesioned rat model of Parkinson's disease. Neurobiol Dis, 70: 138-48. [PMID:24969021]
30. Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. (2004) Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Brain Res Mol Brain Res, 122 (1): 24-34. [PMID:14992813]
31. Lee EK, Ye Y, Kamat AM, Wu X. (2013) Genetic variations in regulator of G-protein signaling (RGS) confer risk of bladder cancer. Cancer, 119 (9): 1643-51. [PMID:23529717]
32. Leontiadis LJ, Papakonstantinou MP, Georgoussi Z. (2009) Regulator of G protein signaling 4 confers selectivity to specific G proteins to modulate mu- and delta-opioid receptor signaling. Cell Signal, 21 (7): 1218-28. [PMID:19324084]
33. Lerner TN, Kreitzer AC. (2012) RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron, 73 (2): 347-59. [PMID:22284188]
34. Liu S, Jiang X, Lu H, Xing M, Qiao Y, Zhang C, Zhang W. (2020) HuR (Human Antigen R) Regulates the Contraction of Vascular Smooth Muscle and Maintains Blood Pressure. Arterioscler Thromb Vasc Biol, 40 (4): 943-957. [PMID:32075416]
35. Liu W, Yuen EY, Allen PB, Feng J, Greengard P, Yan Z. (2006) Adrenergic modulation of NMDA receptors in prefrontal cortex is differentially regulated by RGS proteins and spinophilin. Proc Natl Acad Sci USA, 103 (48): 18338-43. [PMID:17101972]
36. Lur G, Fariborzi M, Higley MJ. (2019) Ketamine disrupts neuromodulatory control of glutamatergic synaptic transmission. PLoS One, 14 (3): e0213721. [PMID:30865708]
37. Madigan LA, Wong GS, Gordon EM, Chen WS, Balenga N, Koziol-White CJ, Panettieri Jr RA, Levine SJ, Druey KM. (2018) RGS4 Overexpression in Lung Attenuates Airway Hyperresponsiveness in Mice. Am J Respir Cell Mol Biol, 58 (1): 89-98. [PMID:28853915]
38. Meng X, Sun X, Zhang Y, Shi H, Deng W, Liu Y, Wang G, Fang P, Yang S. (2018) PPARγ Agonist PGZ Attenuates OVA-Induced Airway Inflammation and Airway Remodeling via RGS4 Signaling in Mouse Model. Inflammation, 41 (6): 2079-2089. [PMID:30022363]
39. Michaelides M, Miller ML, Egervari G, Primeaux SD, Gomez JL, Ellis RJ, Landry JA, Szutorisz H, Hoffman AF, Lupica CR et al.. (2020) Striatal Rgs4 regulates feeding and susceptibility to diet-induced obesity. Mol Psychiatry, 25 (9): 2058-2069. [PMID:29955167]
40. Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. (2001) Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry, 6 (3): 293-301. [PMID:11326297]
41. Monroy CA, Mackie DI, Roman DL. (2013) A high throughput screen for RGS proteins using steady state monitoring of free phosphate formation. PLoS ONE, 8 (4): e62247. [PMID:23626793]
42. Morris DW, Rodgers A, McGhee KA, Schwaiger S, Scully P, Quinn J, Meagher D, Waddington JL, Gill M, Corvin AP. (2004) Confirming RGS4 as a susceptibility gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet, 125B (1): 50-3. [PMID:14755443]
43. Moy FJ, Chanda PK, Cockett MI, Edris W, Jones PG, Mason K, Semus S, Powers R. (2000) NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha. Biochemistry, 39 (24): 7063-73. [PMID:10852703]
44. Opel A, Nobles M, Montaigne D, Finlay M, Anderson N, Breckenridge R, Tinker A. (2015) Absence of the Regulator of G-protein Signaling, RGS4, Predisposes to Atrial Fibrillation and Is Associated with Abnormal Calcium Handling. J Biol Chem, 290 (31): 19233-44. [PMID:26088132]
45. Patella F, Cutler DF. (2020) RGS4 controls secretion of von Willebrand factor to the subendothelial matrix. J Cell Sci, 133 (14). [PMID:32576664]
46. Popov SG, Krishna UM, Falck JR, Wilkie TM. (2000) Ca2+/Calmodulin reverses phosphatidylinositol 3,4, 5-trisphosphate-dependent inhibition of regulators of G protein-signaling GTPase-activating protein activity. J Biol Chem, 275 (25): 18962-8. [PMID:10747990]
47. Roman DL, Blazer LL, Monroy CA, Neubig RR. (2010) Allosteric inhibition of the regulator of G protein signaling-Galpha protein-protein interaction by CCG-4986. Mol Pharmacol, 78 (3): 360-5. [PMID:20530129]
48. Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. (2007) Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol, 71 (1): 169-75. [PMID:17012620]
49. Roof RA, Jin Y, Roman DL, Sunahara RK, Ishii M, Mosberg HI, Neubig RR. (2006) Mechanism of action and structural requirements of constrained peptide inhibitors of RGS proteins. Chem Biol Drug Des, 67 (4): 266-74. [PMID:16629824]
50. Roof RA, Roman DL, Clements ST, Sobczyk-Kojiro K, Blazer LL, Ota S, Mosberg HI, Neubig RR. (2009) A covalent peptide inhibitor of RGS4 identified in a focused one-bead, one compound library screen. BMC Pharmacol, 9: 9. [PMID:19463173]
51. Rorabaugh BR, Rose MJ, Stoops TS, Stevens AA, Seeley SL, D'Souza MS. (2018) Regulators of G-protein signaling 2 and 4 differentially regulate cocaine-induced rewarding effects. Physiol Behav, 195: 9-19. [PMID:30036561]
52. Ruiz de Azua I, Scarselli M, Rosemond E, Gautam D, Jou W, Gavrilova O, Ebert PJ, Levitt P, Wess J. (2010) RGS4 is a negative regulator of insulin release from pancreatic beta-cells in vitro and in vivo. Proc Natl Acad Sci USA, 107 (17): 7999-8004. [PMID:20385802]
53. Siedlecki A, Anderson JR, Jin X, Garbow JR, Lupu TS, Muslin AJ. (2010) RGS4 controls renal blood flow and inhibits cyclosporine-mediated nephrotoxicity. Am J Transplant, 10 (2): 231-41. [PMID:19958325]
54. Sjögren B, Parra S, Heath LJ, Atkins KB, Xie ZJ, Neubig RR. (2012) Cardiotonic steroids stabilize regulator of G protein signaling 2 protein levels. Mol Pharmacol, 82 (3): 500-9. [PMID:22695717]
55. Song KS, Choi YH, Kim JM, Lee H, Lee TJ, Yoon JH. (2009) Suppression of prostaglandin E2-induced MUC5AC overproduction by RGS4 in the airway. Am J Physiol Lung Cell Mol Physiol, 296 (4): L684-92. [PMID:19201815]
56. Storaska AJ, Mei JP, Wu M, Li M, Wade SM, Blazer LL, Sjögren B, Hopkins CR, Lindsley CW, Lin Z et al.. (2013) Reversible inhibitors of regulators of G-protein signaling identified in a high-throughput cell-based calcium signaling assay. Cell Signal, 25 (12): 2848-55. [PMID:24041654]
57. Sullivan BM, Harrison-Lavoie KJ, Marshansky V, Lin HY, Kehrl JH, Ausiello DA, Brown D, Druey KM. (2000) RGS4 and RGS2 bind coatomer and inhibit COPI association with Golgi membranes and intracellular transport. Mol Biol Cell, 11 (9): 3155-68. [PMID:10982407]
58. Tamirisa P, Blumer KJ, Muslin AJ. (1999) RGS4 inhibits G-protein signaling in cardiomyocytes. Circulation, 99 (3): 441-7. [PMID:9918533]
59. Turner EM, Blazer LL, Neubig RR, Husbands SM. (2012) Small Molecule Inhibitors of Regulator of G Protein Signalling (RGS) Proteins. ACS Med Chem Lett, 3 (2): 146-150. [PMID:22368763]
60. Wang HY, MacDonald ML, Borgmann-Winter KE, Banerjee A, Sleiman P, Tom A, Khan A, Lee KC, Roussos P, Siegel SJ et al.. (2020) mGluR5 hypofunction is integral to glutamatergic dysregulation in schizophrenia. Mol Psychiatry, 25 (4): 750-760 RETRACTED. [PMID:30214040]
61. Wang X, Zeng W, Soyombo AA, Tang W, Ross EM, Barnes AP, Milgram SL, Penninger JM, Allen PB, Greengard P et al.. (2005) Spinophilin regulates Ca2+ signalling by binding the N-terminal domain of RGS2 and the third intracellular loop of G-protein-coupled receptors. Nat Cell Biol, 7 (4): 405-11. [PMID:15793568]
62. Wong GS, Redes JL, Balenga N, McCullough M, Fuentes N, Gokhale A, Koziol-White C, Jude JA, Madigan LA, Chan EC et al.. (2020) RGS4 promotes allergen- and aspirin-associated airway hyperresponsiveness by inhibiting PGE2 biosynthesis. J Allergy Clin Immunol, 146 (5): 1152-1164.e13. [PMID:32199913]
63. Xie Y, Wolff DW, Wei T, Wang B, Deng C, Kirui JK, Jiang H, Qin J, Abel PW, Tu Y. (2009) Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res, 69 (14): 5743-51. [PMID:19549919]
64. Xu FL, Yao J, Wu X, Xia X, Xing JX, Xuan JF, Liu YP, Wang BJ. (2020) Association Analysis Between SNPs in the Promoter Region of RGS4 and Schizophrenia in the Northern Chinese Han Population. Neuropsychiatr Dis Treat, 16: 985-992. [PMID:32346293]
65. Zielinski T, Kimple AJ, Hutsell SQ, Koeff MD, Siderovski DP, Lowery RG. (2009) Two Galpha(i1) rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screen, 14 (10): 1195-206. [PMID:19820068]