GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Natural/Endogenous Ligands
- Agonists
- Antagonists
- Immunopharmacology Comments
- Transduction Mechanisms
- Tissue Distribution
- Expression Datasets
- Physiological Functions
- Physiological Consequences of Altering Gene Expression
- Phenotypes, Alleles and Disease Models
- Gene Expression and Pathophysiology
- General Comments
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
||||||
| class A G protein-coupled receptor | ||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 7 | 334 | 3q25.1 | SUCNR1 | succinate receptor 1 | 27 |
| Mouse | 7 | 317 | 3 D | Sucnr1 | succinate receptor 1 | |
| Rat | 7 | 317 | 2q31 | Sucnr1 | succinate receptor 1 | |
| Gene and Protein Information Comments | ||||||
| In humans, there is the possibility of two open-reading frames (ORFs) for SUCNR1, one giving a protein of 330 amino acids (aa) and the other one 334-aa. Wittenberger et al. [27] noted that the 330-aa protein was more likely to be expressed given the Kozak sequence surrounding the second ATG. Some databases report SUCNR1 as being 334-aa long. | ||||||
Previous and Unofficial Names  |
| succinate receptor 1 | GPR91 | P2Y purinoceptor 1 | SUCNR1 | G protein-coupled receptor 91 |
Database Links  |
|
| Specialist databases | |
| GPCRdb | sucr1_human (Hs), sucr1_mouse (Mm), sucr1_rat (Rn) |
| Other databases | |
| Alphafold | Q9BXA5 (Hs), Q99MT6 (Mm), Q6IYF9 (Rn) |
| ChEMBL Target | CHEMBL2150838 (Hs), CHEMBL4739861 (Mm), CHEMBL2150839 (Rn) |
| Ensembl Gene | ENSG00000198829 (Hs), ENSMUSG00000027762 (Mm), ENSRNOG00000014039 (Rn) |
| Entrez Gene | 56670 (Hs), 84112 (Mm), 408199 (Rn) |
| Human Protein Atlas | ENSG00000198829 (Hs) |
| KEGG Gene | hsa:56670 (Hs), mmu:84112 (Mm), rno:408199 (Rn) |
| OMIM | 606381 (Hs) |
| Pharos | Q9BXA5 (Hs) |
| RefSeq Nucleotide | NM_033050 (Hs), NM_032400 (Mm), NM_001001518 (Rn) |
| RefSeq Protein | NP_149039 (Hs), NP_115776 (Mm), NP_001001518 (Rn) |
| UniProtKB | Q9BXA5 (Hs), Q99MT6 (Mm), Q6IYF9 (Rn) |
| Wikipedia | SUCNR1 (Hs) |
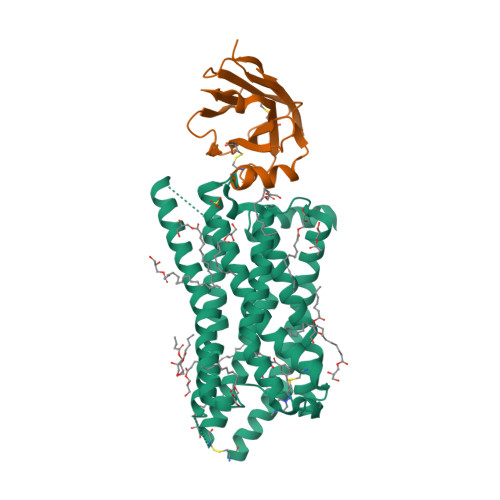
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
Natural/Endogenous Ligands  |
| succinic acid |
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Agonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Using molecular docking calculations, it is predicted that SUCNR1 may interact with gamma-hydroxybutyrate [16]. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific antagonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| There are several synthetic antagonists against SUCNR1 discovered from a systematic structure-activity relationship study [3]. Among these antagonists, N-[{3-(3-trifluoromethyl-4-fluorophenyl)isoxazol-5-yl}methyl]-4-([1,8]naphthyridin-2-yl)butyramide (5g) and 2-{4-[5-(3-Chloro-4-trifluoromethoxyphenyl)-[1,3,4]oxadiazol-2-yl]butyl}-[1,8]naphthyridine (7e) are identified to have high pharmaceutical and clinical importance. 5g demonstrated satisfactory oral bioavailability in rats (%F: 26), very good plasma concentration (Cmax: 37uM, AUC0-24h: 69uM at 30mg/kg p.o. dose) and low plasma clearance. Similarly, 7e demonstrates low plasma clearance (2.0mL/min/kg), excellent bioavailability (%F: 87) and drug exposure (Cmax: 72uM, AUC0-24h: 471uMh, t1/2: 3.4h) upon oral admistration (30mg/kg p.o.) in rats. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Immunopharmacology Comments |
| Succinate acts an an alarmin that triggers the initiation and propagation of danger signals resulting from tissue injury or inflammatory stimuli. It acts through the succinate receptor, SUCNR1. SUCNR1-expressing macrophages release succinate that acts in an autocrine and paracrine feed-forward loop that elevates SUCNR1 expression and leads to enhanced IL-1β production [14]. This mechanism appears to be involved in promoting allergic and autoimmune diseases [20] such as rheumatoid arthritis (RA) (note that succinate levels are elevated in synovial fluids from RA patients), and could potentially be targeted by succinate receptor antagonists for clinical application. |
Primary Transduction Mechanisms 
|
|
| Transducer | Effector/Response |
| Gi/Go family |
Adenylyl cyclase inhibition Phospholipase C stimulation Other - See Comments |
| Comments: The Succinate receptor can recruit arrestins [7] and is internalized upon stimulation [7,11]. The precise mechanism for internalization has not been elucidated. | |
| References: 7,11,23 | |
Secondary Transduction Mechanisms  |
|
| Transducer | Effector/Response |
| Gq/G11 family | Phospholipase C stimulation |
| Comments: Several authors were unable to confirm the activation of the Gq/G11 in transfected HEK293 cells [7,23]. It has been suggested that the Phospholipase C-β activation by Succinate receptor was mediated by Gβγ [23]. | |
| References: 11 | |
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
| Tissue Distribution Comments | ||||||||
| No transcripts of SUCNR1 were detected in the brain when examined by Northern blot analysis [4]. SUCNR1 is functionally expressed by immature dendritic cells after differentiation from monocytes and was rapidly downregulated after dendritic cell maturation [19]. SUCNR1 expression is enriched in both early and late-stage haematopoietic progenitor cells [10]. | ||||||||
Expression Datasets  |
|
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
| Physiological Functions Comments | ||||||||
| SUCNR1 signaling may have a role in activating hepatic stellate cells to restore damaged tissue in the ischemic liver [2]. | ||||||||
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Expression and Pathophysiology Comments | |
| The signalling of SUCNR1 is involved in various pathologies, such as renal hypertension, diabetic nephropathy, retinal angiogenesis, diabetic retinopathy and aggregation of platelets [2-3]. SUCNR1 signalling may also contribue to the formation of fibrosis [2]. |
| General Comments |
| Succinate is as an intermediary of the citric acid cycle. It exerts biological effects via activation of the membrane-bound succinate receptor, SUCNR1 [2]. SUCRN1 is a novel potential drug target to prevent renal complications of diabetes [24]. The expression of SUCNR1 in retinal pigment epithelium cells is iron-dependent [8]. |
References
1. Aguiar CJ, Andrade VL, Gomes ER, Alves MN, Ladeira MS, Pinheiro AC, Gomes DA, Almeida AP, Goes AM, Resende RR et al.. (2010) Succinate modulates Ca(2+) transient and cardiomyocyte viability through PKA-dependent pathway. Cell Calcium, 47 (1): 37-46. [PMID:20018372]
2. Ariza AC, Deen PM, Robben JH. (2012) The succinate receptor as a novel therapeutic target for oxidative and metabolic stress-related conditions. Front Endocrinol (Lausanne), 3: 22. [PMID:22649411]
3. Bhuniya D, Umrani D, Dave B, Salunke D, Kukreja G, Gundu J, Naykodi M, Shaikh NS, Shitole P, Kurhade S et al.. (2011) Discovery of a potent and selective small molecule hGPR91 antagonist. Bioorg Med Chem Lett, 21 (12): 3596-602. [PMID:21571530]
4. Cantagrel V, Lossi AM, Boulanger S, Depetris D, Mattei MG, Gecz J, Schwartz CE, Van Maldergem L, Villard L. (2004) Disruption of a new X linked gene highly expressed in brain in a family with two mentally retarded males. J Med Genet, 41 (10): 736-42. [PMID:15466006]
5. Correa PR, Kruglov EA, Thompson M, Leite MF, Dranoff JA, Nathanson MH. (2007) Succinate is a paracrine signal for liver damage. J Hepatol, 47 (2): 262-9. [PMID:17451837]
6. Geubelle P, Gilissen J, Dilly S, Poma L, Dupuis N, Laschet C, Abboud D, Inoue A, Jouret F, Pirotte B et al.. (2017) Identification and pharmacological characterization of succinate receptor agonists. Br J Pharmacol, 174 (9): 796-808. [PMID:28160606]
7. Gilissen J, Geubelle P, Dupuis N, Laschet C, Pirotte B, Hanson J. (2015) Forskolin-free cAMP assay for Gi-coupled receptors. Biochem Pharmacol, 98 (3): 381-91. [PMID:26386312]
8. Gnana-Prakasam JP, Ananth S, Prasad PD, Zhang M, Atherton SS, Martin PM, Smith SB, Ganapathy V. (2011) Expression and iron-dependent regulation of succinate receptor GPR91 in retinal pigment epithelium. Invest Ophthalmol Vis Sci, 52 (6): 3751-8. [PMID:21357408]
9. Haffke M, Fehlmann D, Rummel G, Boivineau J, Duckely M, Gommermann N, Cotesta S, Sirockin F, Freuler F, Littlewood-Evans A et al.. (2019) Structural basis of species-selective antagonist binding to the succinate receptor. Nature, 574 (7779): 581-585. DOI: 10.1038/s41586-019-1663-8 [PMID:31645725]
10. Hakak Y, Lehmann-Bruinsma K, Phillips S, Le T, Liaw C, Connolly DT, Behan DP. (2009) The role of the GPR91 ligand succinate in hematopoiesis. J Leukoc Biol, 85 (5): 837-43. [PMID:19204147]
11. He W, Miao FJ, Lin DC, Schwandner RT, Wang Z, Gao J, Chen JL, Tian H, Ling L. (2004) Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature, 429 (6988): 188-93. [PMID:15141213]
12. Hebert SC. (2004) Physiology: orphan detectors of metabolism. Nature, 429 (6988): 143-5. [PMID:15141197]
13. Högberg C, Gidlöf O, Tan C, Svensson S, Nilsson-Öhman J, Erlinge D, Olde B. (2011) Succinate independently stimulates full platelet activation via cAMP and phosphoinositide 3-kinase-β signaling. J Thromb Haemost, 9 (2): 361-72. [PMID:21143371]
14. Littlewood-Evans A, Sarret S, Apfel V, Loesle P, Dawson J, Zhang J, Muller A, Tigani B, Kneuer R, Patel S et al.. (2016) GPR91 senses extracellular succinate released from inflammatory macrophages and exacerbates rheumatoid arthritis. J Exp Med, 213 (9): 1655-62. [PMID:27481132]
15. Macaulay IC, Tijssen MR, Thijssen-Timmer DC, Gusnanto A, Steward M, Burns P, Langford CF, Ellis PD, Dudbridge F, Zwaginga JJ, Watkins NA, van der Schoot CE, Ouwehand WH. (2007) Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood, 109 (8): 3260-9. [PMID:17192395]
16. Molnár T, Héja L, Emri Z, Simon A, Nyitrai G, Pál I, Kardos J. (2011) Activation of astroglial calcium signaling by endogenous metabolites succinate and gamma-hydroxybutyrate in the nucleus accumbens. Front Neuroenergetics, 3: 7. [PMID:22180742]
17. Regard JB, Sato IT, Coughlin SR. (2008) Anatomical profiling of G protein-coupled receptor expression. Cell, 135 (3): 561-71. [PMID:18984166]
18. Rexen Ulven E, Trauelsen M, Brvar M, Lückmann M, Bielefeldt LØ, Jensen LKI, Schwartz TW, Frimurer TM. (2018) Structure-Activity Investigations and Optimisations of Non-metabolite Agonists for the Succinate Receptor 1. Sci Rep, 8 (1): 10010. [PMID:29968758]
19. Rubic T, Lametschwandtner G, Jost S, Hinteregger S, Kund J, Carballido-Perrig N, Schwärzler C, Junt T, Voshol H, Meingassner JG et al.. (2008) Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat Immunol, 9 (11): 1261-9. [PMID:18820681]
20. Rubić-Schneider T, Carballido-Perrig N, Regairaz C, Raad L, Jost S, Rauld C, Christen B, Wieczorek G, Kreutzer R, Dawson J et al.. (2017) GPR91 deficiency exacerbates allergic contact dermatitis while reducing arthritic disease in mice. Allergy, 72 (3): 444-452. [PMID:27527650]
21. Sapieha P, Sirinyan M, Hamel D, Zaniolo K, Joyal JS, Cho JH, Honoré JC, Kermorvant-Duchemin E, Varma DR, Tremblay S et al.. (2008) The succinate receptor GPR91 in neurons has a major role in retinal angiogenesis. Nat Med, 14 (10): 1067-76. [PMID:18836459]
22. Southern C, Cook JM, Neetoo-Isseljee Z, Taylor DL, Kettleborough CA, Merritt A, Bassoni DL, Raab WJ, Quinn E, Wehrman TS et al.. (2013) Screening β-Arrestin Recruitment for the Identification of Natural Ligands for Orphan G-Protein-Coupled Receptors. J Biomol Screen, 18 (5): 599-609. [PMID:23396314]
23. Sundström L, Greasley PJ, Engberg S, Wallander M, Ryberg E. (2013) Succinate receptor GPR91, a Gα(i) coupled receptor that increases intracellular calcium concentrations through PLCβ. FEBS Lett, 587 (15): 2399-404. [PMID:23770096]
24. Toma I, Kang JJ, Sipos A, Vargas S, Bansal E, Hanner F, Meer E, Peti-Peterdi J. (2008) Succinate receptor GPR91 provides a direct link between high glucose levels and renin release in murine and rabbit kidney. J Clin Invest, 118 (7): 2526-34. [PMID:18535668]
25. Trauelsen M, Rexen Ulven E, Hjorth SA, Brvar M, Monaco C, Frimurer TM, Schwartz TW. (2017) Receptor structure-based discovery of non-metabolite agonists for the succinate receptor GPR91. Mol Metab, 6 (12): 1585-1596. [PMID:29157600]
26. Vargas SL, Toma I, Kang JJ, Meer EJ, Peti-Peterdi J. (2009) Activation of the succinate receptor GPR91 in macula densa cells causes renin release. J Am Soc Nephrol, 20 (5): 1002-11. [PMID:19389848]
27. Wittenberger T, Schaller HC, Hellebrand S. (2001) An expressed sequence tag (EST) data mining strategy succeeding in the discovery of new G-protein coupled receptors. J Mol Biol, 307 (3): 799-813. [PMID:11273702]