GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
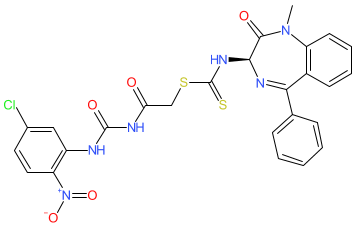
Synonyms: (+)-SL18
Compound class:
Synthetic organic
Comment: (R)-SL18 is reported as a small molecule degrader of annexin A3 (ANXA3) protein, with degradation mediated principally via the ubiquitin-proteasome system (UPS) [1]. It was superseded by ANXA3 degrader 18a5 which has optimised selectivity and improved binding affinity and anti-proliferative effects in target TNBC cancer cells.
|
|
|||||||||||||||||||||||||||||||||||
| Bioactivity Comments |
| The DC50 for ANXA3 protein in TNBC cell lines is ~2-3 μM [1]. (R)-SL18 can bind to additional ANXA proteins in vitro, but protein degradation was only detectable for ANXA4 and ANXA6. |
| Selectivity at ligand targets | ||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||