GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
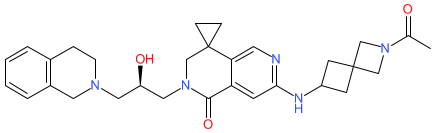
Synonyms: compound D3 [PMID: 39722476]
Compound class:
Synthetic organic
Comment: This small molecule behaves as a selective inhibitor of protein arginine methyltransferase 5 (PRMT5) [1]. Its structure was designed through scaffold-hopping and SAR based on the non-nucleoside ex-clinical lead inhibitor GSK3326595.
Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖ |
|
|||||||||||||||||||||||||||||||||||
| Bioactivity Comments |
| D3 has demonstrated anti-tumour activity in vitro and in vivo [1]. It induces cell cycle arrest and apoptosis in lymphoma models, and is selective for PRMT5 compared to other protein arginine N-methyltransferases. |
| Selectivity at enzymes | ||||||||||||||||||||||||||||||||||
| Key to terms and symbols | Click column headers to sort | |||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||