GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
Synonyms: Compound A [WO2021257532A1] | example 34 [US20210155606A1] | INCB-00928 | INCB00928
Compound class:
Synthetic organic
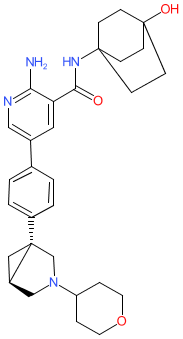
Comment: The chemical structure for zilurgisertib was revealed in WHO proposed INN list 126 (Jan 2022), in which it was described as a serine/threonine kinase inhibitor. It is one of the compounds that are claimed in Novartis' ALK2 (ACVR1) inhibitor patent US20210155606A1 [1], and it is also claimed in WO2021257532A1 by Incyte as "Compound A" [2]. Incyte have declared an ALK2 inhibitor (INCB00928) in their development pipeline. Name-to-structure was formally disclosed in early 2025 [3]. Zilurgisertib (INCB00928) is declared as an ATP-competitive, orally available and selective ALK2/ACVR1 inhibitor.
ALK2/ACVR1 is a serine/threonine protein kinase receptor for type I bone morphogenic protein (BMP), and this signalling pathway is crucial in bone formation, and hyperactivty of the pathway (e.g. by gain-of-function mutation ALK2R206H) is associated with diseases that are characterised by bone malformation, such as fibrodysplasia ossificans progressiva (FOP). ALK2 is also involved in the maintenance of iron homeostasis by governing production of the iron regulatory hormone hepcidin. Elevated hepcidin is thought to be a powerful driver of the pathophysiology of anemia that's associated with chronic disease. Inhibition of ALK2 upstream of hepcidin should increase circulating iron levels. Thus direct inhibition of ALK2/ACVR1 is a molecular mechanism that is being exploited for therapeutic potential in FOP and anemia arising from chronic kidney disease or myeloproliferative diseases. Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖ |
|
|||||||||||||||||||||||||||||||||||
Download 2D Structure  |
|
| Canonical SMILES | Download |
| Isomeric SMILES | Download |
| InChI standard identifier | Download |
| InChI standard key | Download |
Molecular structure representations generated using Open Babel