GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Contents:
Gene and Protein Information  |
||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | - | 192 | 1q21.3 | PMVK | phosphomevalonate kinase | 5 |
| Mouse | - | 192 | 3 F1 | Pmvk | phosphomevalonate kinase | |
| Rat | - | 194 | 2q34 | Pmvk | phosphomevalonate kinase | |
Previous and Unofficial Names  |
| ATP:5-phosphomevalonate phosphotransferase | mevalonate-5-phosphate kinase | mevalonic acid phosphate kinase | phosphomevalonic kinase | PMK | HUMPMKI | PMKA |
Database Links  |
|
| Alphafold | Q15126 (Hs), Q9D1G2 (Mm), Q5RK24 (Rn) |
| BRENDA | 2.7.4.2 |
| Ensembl Gene | ENSG00000163344 (Hs), ENSMUSG00000027952 (Mm), ENSRNOG00000020696 (Rn) |
| Entrez Gene | 10654 (Hs), 68603 (Mm), 310645 (Rn) |
| Human Protein Atlas | ENSG00000163344 (Hs) |
| KEGG Enzyme | 2.7.4.2 |
| KEGG Gene | hsa:10654 (Hs), mmu:68603 (Mm), rno:310645 (Rn) |
| OMIM | 607622 (Hs) |
| Pharos | Q15126 (Hs) |
| RefSeq Nucleotide | NM_006556 (Hs), NM_026784 (Mm), NM_027348 (Mm), NM_001008352 (Rn) |
| RefSeq Protein | NP_006547 (Hs), NP_081060 (Mm), NP_081624 (Mm), NP_001008353 (Rn) |
| UniProtKB | Q15126 (Hs), Q9D1G2 (Mm), Q5RK24 (Rn) |
| Wikipedia | PMVK (Hs) |
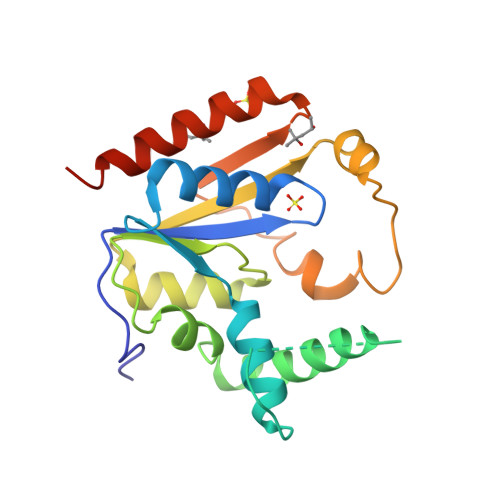
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
Enzyme Reaction  |
||||
|
||||
Substrates and Reaction Kinetics  |
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Cofactors  |
||||||||
|
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Tissue Distribution 
|
||||||||||
|
||||||||||
|
References
1. Chambliss KL, Slaughter CA, Schreiner R, Hoffmann GF, Gibson KM. (1996) Molecular cloning of human phosphomevalonate kinase and identification of a consensus peroxisomal targeting sequence. J Biol Chem, 271 (29): 17330-4. [PMID:8663599]
2. Chang Q, Yan XX, Gu SY, Liu JF, Liang DC. (2008) Crystal structure of human phosphomevalonate kinase at 1.8 A resolution. Proteins, 73 (1): 254-8. [PMID:18618710]
3. Herdendorf TJ, Miziorko HM. (2006) Phosphomevalonate kinase: functional investigation of the recombinant human enzyme. Biochemistry, 45 (10): 3235-42. [PMID:16519518]
4. Herdendorf TJ, Miziorko HM. (2007) Functional evaluation of conserved basic residues in human phosphomevalonate kinase. Biochemistry, 46 (42): 11780-8. [PMID:17902708]
5. Olivier LM, Chambliss KL, Gibson KM, Krisans SK. (1999) Characterization of phosphomevalonate kinase: chromosomal localization, regulation, and subcellular targeting. J Lipid Res, 40 (4): 672-9. [PMID:10191291]
6. Popjak G, Parker TS, Sarin V, Tropp BE, Engel R. (1978) Inhibition of 5-phosphomevalonate kinase by an isosteric analog of 5-phosphomevalonate. J Am Chem Soc, 100 (25): 8014-8016.
7. Shama Bhat C, Ramasarma T. (1979) Inhibition of rat liver mevalonate pyrophosphate decarboxylase and mevalonate phosphate kinase by phenyl and phenolic compounds. Biochem J, 181 (1): 143-51. [PMID:226078]