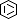GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Natural/Endogenous Ligands
- Rank order lists
- Agonists
- Antagonists
- Allosteric Modulators
- Immunopharmacology Comments
- Transduction Mechanisms
- Expression Datasets
- Physiological Consequences of Altering Gene Expression
- Phenotypes, Alleles and Disease Models
- Biologically Significant Variants
- General Comments
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
||||||
| class A G protein-coupled receptor | ||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 7 | 397 | 5q13.3 | F2RL1 | F2R like trypsin receptor 1 | |
| Mouse | 7 | 399 | 13 49.53 cM | F2rl1 | F2R like trypsin receptor 1 | |
| Rat | 7 | 397 | 2q12 | F2rl1 | F2R like trypsin receptor 1 | |
Previous and Unofficial Names  |
| GPR11 | coagulation factor II receptor-like 1 | Protease-activated receptor-2 | coagulation factor II (thrombin) receptor-like 1 |
Database Links  |
|
| Specialist databases | |
| GPCRdb | par2_human (Hs), par2_mouse (Mm), par2_rat (Rn) |
| Other databases | |
| Alphafold | P55085 (Hs), P55086 (Mm), Q63645 (Rn) |
| ChEMBL Target | CHEMBL5963 (Hs), CHEMBL3734647 (Mm), CHEMBL2429706 (Rn) |
| Ensembl Gene | ENSG00000164251 (Hs), ENSMUSG00000021678 (Mm), ENSRNOG00000018003 (Rn) |
| Entrez Gene | 2150 (Hs), 14063 (Mm), 116677 (Rn) |
| Human Protein Atlas | ENSG00000164251 (Hs) |
| KEGG Gene | hsa:2150 (Hs), mmu:14063 (Mm), rno:116677 (Rn) |
| OMIM | 600933 (Hs) |
| Pharos | P55085 (Hs) |
| RefSeq Nucleotide | NM_005242 (Hs), NM_007974 (Mm), NM_053897 (Rn) |
| RefSeq Protein | NP_005233 (Hs), NP_032000 (Mm), NP_446349 (Rn) |
| SynPHARM |
84335 (in complex with AZ3451) 84336 (in complex with AZ3451) 84445 (in complex with AZ3451) 84449 (in complex with AZ8838) 84447 (in complex with AZ8838) 84448 (in complex with AZ8838) |
| UniProtKB | P55085 (Hs), P55086 (Mm), Q63645 (Rn) |
| Wikipedia | F2RL1 (Hs) |
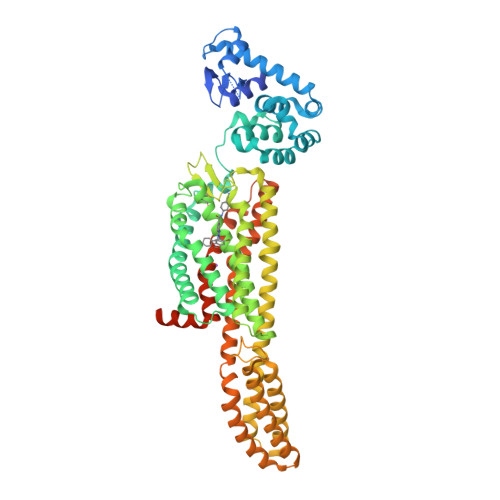
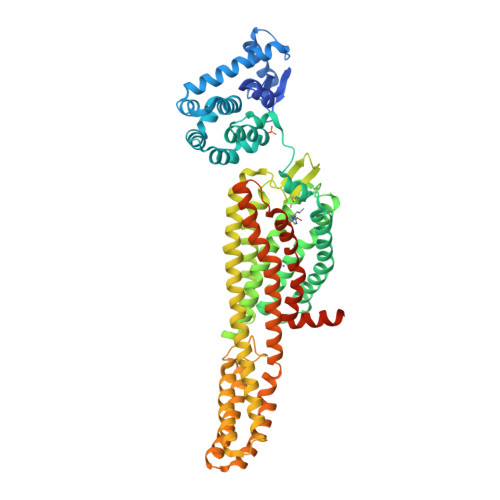
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
Natural/Endogenous Ligands  |
| serine proteases |
| Agonist proteases (Human) |
| Trypsin, tryptase, TF/VIIa, Xa; elastase, neutrophil expressed; cathepsin S [21,46] |
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Agonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2-Furoyl-LIGRLO-NH2 activity was measured via calcium mobilisation in HEK 293 cells which constitutively coexpress human PAR1 and PAR2. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulators | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Immunopharmacology Comments |
| PAR2 receptors have been reported to elicit pain and inflammation through a neurogenic mechanism of action, causing release of substance P, activation of NK1 receptors, and sensitization of TRPV1 voltage-gated ion channels. This action can be negated using a selective NK1 receptor antagonist (L732,138) or a TRPV1 receptor antagonist (capsazepine) [15]. PAR2 receptors are expressed in immune cells of both the innate and adaptive immune systems and have been shown to play a role in several peripheral inflammatory conditions. PAR2 activation has been associated with allergic airway inflammation, and allergic sensitisation. Anti-PAR2 antibody or PAR2 inhibiting pepducin alleviate allergen-induced airway hyperresponsiveness and inflammation in mice [35]. PAR2 expression by astrocytes and microglia within the CNS suggests that PAR2 modulation may be a novel approach for developing therapeutics for CNS disorders with an inflammatory association [7]. A study in obese mice suggests that activation of PAR2 is associated with increased adipose expression of inflammatory factors [35]. |
Primary Transduction Mechanisms 
|
|
| Transducer | Effector/Response |
|
Gi/Go family Gq/G11 family G12/G13 family |
|
| References: | |
Expression Datasets  |
|
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Biologically Significant Variants 
|
||||||||||||
|
||||||||||||
|
| General Comments |
|
SARS-CoV-2/COVID-19 Experimental in vitro evidence, using affinity-purification mass spectrometry (AP-MS), indicates a protein-protein interaction between PAR2 and the SARS-CoV-2 Orf9c protein [17], although whether this interaction is realistic based on spatial distribution of the host and viral proteins within cells was not addressed in this study. Speculatively, PAR2 ligands such as AZ3451, GB110, or AZ8838 could be utilised to examine the effect of blocking the PAR2/Orf9c protein-protein interaction on SARS-CoV-2 pathobiology. |
References
1. Al-Ani B, Saifeddine M, Kawabata A, Renaux B, Mokashi S, Hollenberg MD. (1999) Proteinase-activated receptor 2 (PAR(2)): development of a ligand-binding assay correlating with activation of PAR(2) by PAR(1)- and PAR(2)-derived peptide ligands. J Pharmacol Exp Ther, 290 (2): 753-60. [PMID:10411588]
2. Amiable N, Martel-Pelletier J, Lussier B, Kwan Tat S, Pelletier JP, Boileau C. (2011) Proteinase-activated receptor-2 gene disruption limits the effect of osteoarthritis on cartilage in mice: a novel target in joint degradation. J Rheumatol, 38 (5): 911-20. [PMID:21285164]
3. Avet C, Sturino C, Grastilleur S, Gouill CL, Semache M, Gross F, Gendron L, Bennani Y, Mancini JA, Sayegh CE et al.. (2020) The PAR2 inhibitor I-287 selectively targets Gαq and Gα12/13 signaling and has anti-inflammatory effects. Commun Biol, 3 (1): 719. [PMID:33247181]
4. Badeanlou L, Furlan-Freguia C, Yang G, Ruf W, Samad F. (2011) Tissue factor-protease-activated receptor 2 signaling promotes diet-induced obesity and adipose inflammation. Nat Med, 17 (11): 1490-7. [PMID:22019885]
5. Barry GD, Suen JY, Le GT, Cotterell A, Reid RC, Fairlie DP. (2010) Novel agonists and antagonists for human protease activated receptor 2. J Med Chem, 53 (20): 7428-40. [PMID:20873792]
6. Boitano S, Hoffman J, Flynn AN, Asiedu MN, Tillu DV, Zhang Z, Sherwood CL, Rivas CM, DeFea KA, Vagner J et al.. (2015) The novel PAR2 ligand C391 blocks multiple PAR2 signalling pathways in vitro and in vivo. Br J Pharmacol, 172 (18): 4535-4545. [PMID:26140338]
7. Bushell TJ, Cunningham MR, McIntosh KA, Moudio S, Plevin R. (2016) Protease-Activated Receptor 2: Are Common Functions in Glial and Immune Cells Linked to Inflammation-Related CNS Disorders?. Curr Drug Targets, 17 (16): 1861-1870. [PMID:26648078]
8. Busso N, Frasnelli M, Feifel R, Cenni B, Steinhoff M, Hamilton J, So A. (2007) Evaluation of protease-activated receptor 2 in murine models of arthritis. Arthritis Rheum, 56 (1): 101-7. [PMID:17195212]
9. Cheng RKY, Fiez-Vandal C, Schlenker O, Edman K, Aggeler B, Brown DG, Brown GA, Cooke RM, Dumelin CE, Doré AS et al.. (2017) Structural insight into allosteric modulation of protease-activated receptor 2. Nature, 545 (7652): 112-115. [PMID:28445455]
10. Cottrell GS, Amadesi S, Pikios S, Camerer E, Willardsen JA, Murphy BR, Caughey GH, Wolters PJ, Coughlin SR, Peterson A et al.. (2007) Protease-activated receptor 2, dipeptidyl peptidase I, and proteases mediate Clostridium difficile toxin A enteritis. Gastroenterology, 132 (7): 2422-37. [PMID:17570216]
11. Crilly A, Palmer H, Nickdel MB, Dunning L, Lockhart JC, Plevin R, McInnes IB, Ferrell WR. (2012) Immunomodulatory role of proteinase-activated receptor-2. Ann Rheum Dis, 71 (9): 1559-66. [PMID:22563031]
12. Davidson CE, Asaduzzaman M, Arizmendi NG, Polley D, Wu Y, Gordon JR, Hollenberg MD, Cameron L, Vliagoftis H. (2013) Proteinase-activated receptor-2 activation participates in allergic sensitization to house dust mite allergens in a murine model. Clin Exp Allergy, 43 (11): 1274-85. [PMID:24152160]
13. de Boer JD, Van't Veer C, Stroo I, van der Meer AJ, de Vos AF, van der Zee JS, Roelofs JJ, van der Poll T. (2014) Protease-activated receptor-2 deficient mice have reduced house dust mite-evoked allergic lung inflammation. Innate Immun, 20 (6): 618-25. [PMID:24048772]
14. De Cunto G, Cardini S, Cirino G, Geppetti P, Lungarella G, Lucattelli M. (2011) Pulmonary hypertension in smoking mice over-expressing protease-activated receptor-2. Eur Respir J, 37 (4): 823-34. [PMID:20693251]
15. Gardell LR, Ma JN, Seitzberg JG, Knapp AE, Schiffer HH, Tabatabaei A, Davis CN, Owens M, Clemons B, Wong KK et al.. (2008) Identification and characterization of novel small-molecule protease-activated receptor 2 agonists. J Pharmacol Exp Ther, 327 (3): 799-808. [PMID:18768780]
16. Georgy SR, Pagel CN, Ghasem-Zadeh A, Zebaze RM, Pike RN, Sims NA, Mackie EJ. (2012) Proteinase-activated receptor-2 is required for normal osteoblast and osteoclast differentiation during skeletal growth and repair. Bone, 50 (3): 704-12. [PMID:22173052]
17. Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL et al.. (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature, 583 (7816): 459-468. [PMID:32353859]
18. Hollenberg MD, Renaux B, Hyun E, Houle S, Vergnolle N, Saifeddine M, Ramachandran R. (2008) Derivatized 2-furoyl-LIGRLO-amide, a versatile and selective probe for proteinase-activated receptor 2: binding and visualization. J Pharmacol Exp Ther, 326 (2): 453-62. [PMID:18477767]
19. Holzhausen M, Spolidorio LC, Ellen RP, Jobin MC, Steinhoff M, Andrade-Gordon P, Vergnolle N. (2006) Protease-activated receptor-2 activation: a major role in the pathogenesis of Porphyromonas gingivalis infection. Am J Pathol, 168 (4): 1189-99. [PMID:16565494]
20. Jiang Y, Yau MK, Lim J, Wu KC, Xu W, Suen JY, Fairlie DP. (2018) A Potent Antagonist of Protease-Activated Receptor 2 That Inhibits Multiple Signaling Functions in Human Cancer Cells. J Pharmacol Exp Ther, 364 (2): 246-257. [PMID:29263243]
21. Jimenez-Vargas NN, Pattison LA, Zhao P, Lieu T, Latorre R, Jensen DD, Castro J, Aurelio L, Le GT, Flynn B et al.. (2018) Protease-activated receptor-2 in endosomes signals persistent pain of irritable bowel syndrome. Proc Natl Acad Sci USA, 115 (31): E7438-E7447. [PMID:30012612]
22. Jin G, Hayashi T, Kawagoe J, Takizawa T, Nagata T, Nagano I, Syoji M, Abe K. (2005) Deficiency of PAR-2 gene increases acute focal ischemic brain injury. J Cereb Blood Flow Metab, 25 (3): 302-13. [PMID:15647743]
23. Kanke T, Ishiwata H, Kabeya M, Saka M, Doi T, Hattori Y, Kawabata A, Plevin R. (2005) Binding of a highly potent protease-activated receptor-2 (PAR2) activating peptide, [3H]2-furoyl-LIGRL-NH2, to human PAR2. Br J Pharmacol, 145 (2): 255-63. [PMID:15765104]
24. Kawabata A, Matsunami M, Tsutsumi M, Ishiki T, Fukushima O, Sekiguchi F, Kawao N, Minami T, Kanke T, Saito N. (2006) Suppression of pancreatitis-related allodynia/hyperalgesia by proteinase-activated receptor-2 in mice. Br J Pharmacol, 148 (1): 54-60. [PMID:16520745]
25. Kawagoe J, Takizawa T, Matsumoto J, Tamiya M, Meek SE, Smith AJ, Hunter GD, Plevin R, Saito N, Kanke T et al.. (2002) Effect of protease-activated receptor-2 deficiency on allergic dermatitis in the mouse ear. Jpn J Pharmacol, 88 (1): 77-84. [PMID:11859856]
26. Kennedy AJ, Sundström L, Geschwindner S, Poon EKY, Jiang Y, Chen R, Cooke R, Johnstone S, Madin A, Lim J et al.. (2020) Protease-activated receptor-2 ligands reveal orthosteric and allosteric mechanisms of receptor inhibition. Commun Biol, 3 (1): 782. [PMID:33335291]
27. Khoufache K, LeBouder F, Morello E, Laurent F, Riffault S, Andrade-Gordon P, Boullier S, Rousset P, Vergnolle N, Riteau B. (2009) Protective role for protease-activated receptor-2 against influenza virus pathogenesis via an IFN-gamma-dependent pathway. J Immunol, 182 (12): 7795-802. [PMID:19494303]
28. Knight V, Tchongue J, Lourensz D, Tipping P, Sievert W. (2012) Protease-activated receptor 2 promotes experimental liver fibrosis in mice and activates human hepatic stellate cells. Hepatology, 55 (3): 879-87. [PMID:22095855]
29. Lam DK, Schmidt BL. (2010) Serine proteases and protease-activated receptor 2-dependent allodynia: a novel cancer pain pathway. Pain, 149 (2): 263-72. [PMID:20189717]
30. Lee JH, Kim KW, Gee HY, Lee J, Lee KH, Park HS, Kim SH, Kim SW, Kim MN, Kim KE et al.. (2011) A synonymous variation in protease-activated receptor-2 is associated with atopy in Korean children. J Allergy Clin Immunol, 128 (6): 1326-1334.e3. [PMID:21839502]
31. Lee MC, Huang SC. (2008) Proteinase-activated receptor-1 (PAR(1)) and PAR(2) but not PAR(4) mediate contraction in human and guinea-pig gallbladders. Neurogastroenterol Motil, 20 (4): 385-91. [PMID:18179608]
32. LeSarge JC, Thibeault P, Milne M, Ramachandran R, Luyt LG. (2019) High Affinity Fluorescent Probe for Proteinase-Activated Receptor 2 (PAR2). ACS Med Chem Lett, 10 (7): 1045-1050. [PMID:31312406]
33. LeSarge JC, Thibeault P, Yu L, Childs MD, Mirka VM, Qi Q, Fox MS, Kovacs MS, Ramachandran R, Luyt LG. (2023) Protease-activated receptor 2 (PAR2)-targeting peptide derivatives for positron emission tomography (PET) imaging. Eur J Med Chem, 246: 114989. [PMID:36527934]
34. Lewkowich IP, Day SB, Ledford JR, Zhou P, Dienger K, Wills-Karp M, Page K. (2011) Protease-activated receptor 2 activation of myeloid dendritic cells regulates allergic airway inflammation. Respir Res, 12: 122. [PMID:21936897]
35. Li M, Yang X, Zhang Y, Chen L, Lu H, Li X, Yin L, Zhi X. (2015) Activation of protease‑activated receptor‑2 is associated with increased expression of inflammatory factors in the adipose tissues of obese mice. Mol Med Rep, 12 (4): 6227-34. [PMID:26252901]
36. Lindner JR, Kahn ML, Coughlin SR, Sambrano GR, Schauble E, Bernstein D, Foy D, Hafezi-Moghadam A, Ley K. (2000) Delayed onset of inflammation in protease-activated receptor-2-deficient mice. J Immunol, 165 (11): 6504-10. [PMID:11086091]
37. Liu S, Liu YP, Yue DM, Liu GJ. (2014) Protease-activated receptor 2 in dorsal root ganglion contributes to peripheral sensitization of bone cancer pain. Eur J Pain, 18 (3): 326-37. [PMID:23893658]
38. Liu Y, Ji W, Yin Y, Fan L, Zhang J, Yun H, Wang N, Li Q, Wei Z, Ouyang D et al.. (2009) The effects of splicing variant of PXR PAR-2 on CYP3A4 and MDR1 mRNA expressions. Clin Chim Acta, 403 (1-2): 142-4. [PMID:19232333]
39. Matej R, Olejar T, Janouskova O, Holada K. (2012) Deletion of protease-activated receptor 2 prolongs survival of scrapie-inoculated mice. J Gen Virol, 93 (Pt 9): 2057-61. [PMID:22694901]
40. McGuire JJ, Saifeddine M, Triggle CR, Sun K, Hollenberg MD. (2004) 2-furoyl-LIGRLO-amide: a potent and selective proteinase-activated receptor 2 agonist. J Pharmacol Exp Ther, 309 (3): 1124-31. [PMID:14976230]
41. McGuire JJ, Van Vliet BN, Halfyard SJ. (2008) Blood pressures, heart rate and locomotor activity during salt loading and angiotensin II infusion in protease-activated receptor 2 (PAR2) knockout mice. BMC Physiol, 8: 20. [PMID:18939990]
42. Moussa L, Apostolopoulos J, Davenport P, Tchongue J, Tipping PG. (2007) Protease-activated receptor-2 augments experimental crescentic glomerulonephritis. Am J Pathol, 171 (3): 800-8. [PMID:17640968]
43. Noorbakhsh F, Tsutsui S, Vergnolle N, Boven LA, Shariat N, Vodjgani M, Warren KG, Andrade-Gordon P, Hollenberg MD, Power C. (2006) Proteinase-activated receptor 2 modulates neuroinflammation in experimental autoimmune encephalomyelitis and multiple sclerosis. J Exp Med, 203 (2): 425-35. [PMID:16476770]
44. Olejar T, Vetvicka D, Zadinova M, Pouckova P, Kukal J, Jezek P, Matej R. (2014) Dual role of host Par2 in a murine model of spontaneous metastatic B16 melanoma. Anticancer Res, 34 (7): 3511-5. [PMID:24982362]
45. Page K, Ledford JR, Zhou P, Dienger K, Wills-Karp M. (2010) Mucosal sensitization to German cockroach involves protease-activated receptor-2. Respir Res, 11: 62. [PMID:20497568]
46. Ramachandran R, Mihara K, Chung H, Renaux B, Lau CS, Muruve DA, DeFea KA, Bouvier M, Hollenberg MD. (2011) Neutrophil elastase acts as a biased agonist for proteinase-activated receptor-2 (PAR2). J Biol Chem, 286 (28): 24638-48. [PMID:21576245]
47. Schaffner F, Versteeg HH, Schillert A, Yokota N, Petersen LC, Mueller BM, Ruf W. (2010) Cooperation of tissue factor cytoplasmic domain and PAR2 signaling in breast cancer development. Blood, 116 (26): 6106-13. [PMID:20861457]
48. Seeliger S, Derian CK, Vergnolle N, Bunnett NW, Nawroth R, Schmelz M, Von Der Weid PY, Buddenkotte J, Sunderkötter C, Metze D et al.. (2003) Proinflammatory role of proteinase-activated receptor-2 in humans and mice during cutaneous inflammation in vivo. FASEB J, 17 (13): 1871-85. [PMID:14519665]
49. Seitzberg JG, Knapp AE, Lund BW, Mandrup Bertozzi S, Currier EA, Ma JN, Sherbukhin V, Burstein ES, Olsson R. (2008) Discovery of potent and selective small-molecule PAR-2 agonists. J Med Chem, 51 (18): 5490-3. [PMID:18720984]
50. Singh VP, Bhagat L, Navina S, Sharif R, Dawra RK, Saluja AK. (2007) Protease-activated receptor-2 protects against pancreatitis by stimulating exocrine secretion. Gut, 56 (7): 958-64. [PMID:17114298]
51. Suen JY, Barry GD, Lohman RJ, Halili MA, Cotterell AJ, Le GT, Fairlie DP. (2012) Modulating human proteinase activated receptor 2 with a novel antagonist (GB88) and agonist (GB110). Br J Pharmacol, 165 (5): 1413-23. [PMID:21806599]
52. Takizawa T, Tamiya M, Hara T, Matsumoto J, Saito N, Kanke T, Kawagoe J, Hattori Y. (2005) Abrogation of bronchial eosinophilic inflammation and attenuated eotaxin content in protease-activated receptor 2-deficient mice. J Pharmacol Sci, 98 (1): 99-102. [PMID:15879675]
53. Uusitalo-Jarvinen H, Kurokawa T, Mueller BM, Andrade-Gordon P, Friedlander M, Ruf W. (2007) Role of protease activated receptor 1 and 2 signaling in hypoxia-induced angiogenesis. Arterioscler Thromb Vasc Biol, 27 (6): 1456-62. [PMID:17363687]
54. Versteeg HH, Schaffner F, Kerver M, Ellies LG, Andrade-Gordon P, Mueller BM, Ruf W. (2008) Protease-activated receptor (PAR) 2, but not PAR1, signaling promotes the development of mammary adenocarcinoma in polyoma middle T mice. Cancer Res, 68 (17): 7219-27. [PMID:18757438]
55. Weithauser A, Bobbert P, Antoniak S, Böhm A, Rauch BH, Klingel K, Savvatis K, Kroemer HK, Tschope C, Stroux A et al.. (2013) Protease-activated receptor-2 regulates the innate immune response to viral infection in a coxsackievirus B3-induced myocarditis. J Am Coll Cardiol, 62 (19): 1737-45. [PMID:23871888]
56. Wong DM, Tam V, Lam R, Walsh KA, Tatarczuch L, Pagel CN, Reynolds EC, O'Brien-Simpson NM, Mackie EJ, Pike RN. (2010) Protease-activated receptor 2 has pivotal roles in cellular mechanisms involved in experimental periodontitis. Infect Immun, 78 (2): 629-38. [PMID:19933835]
57. Xue M, Chan YK, Shen K, Dervish S, March L, Sambrook PN, Jackson CJ. (2012) Protease-activated receptor 2, rather than protease-activated receptor 1, contributes to the aggressive properties of synovial fibroblasts in rheumatoid arthritis. Arthritis Rheum, 64 (1): 88-98. [PMID:21905006]
58. Yau MK, Suen JY, Xu W, Lim J, Liu L, Adams MN, He Y, Hooper JD, Reid RC, Fairlie DP. (2016) Potent Small Agonists of Protease Activated Receptor 2. ACS Med Chem Lett, 7 (1): 105-10. [PMID:26819675]