GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Contents:
Database Links  |
|
| UniProtKB | P0DTD1 (SARS-CoV-2) |
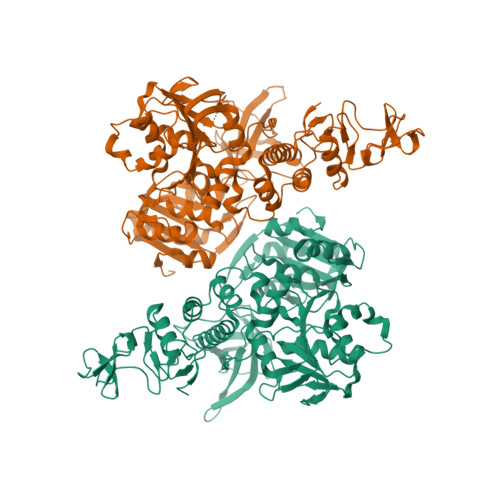
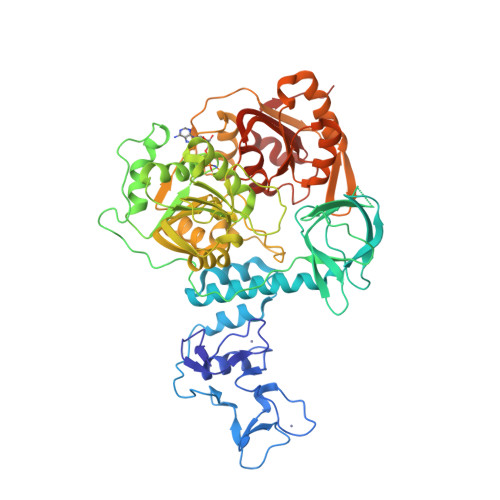
Selected 3D Structures  |
|||||||||||
|
|
||||||||||
|
|
||||||||||
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulators | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| General Comments |
| Non-structural protein 13 is the most highly conserved protein among all known coronaviruses [17-18]. It has helicase unwinding activity (5′-3′: IUBMB enzyme nomenclature identifier EC 5.6.2.3) on DNA and RNA, is a component of the RNA-synthesis complex, and is therefore crucial for viral replication. Small molecule inhibitors of MERS-CoV and SARS-CoV-1 helicases were reported [7-8,19-20]. Renewed interest in developing nsp13 inhibitor-based antivirals arose in response to the SARS-CoV-2 pandemic [5,9-12,14]. Nsp13 is also reported to interfere with the host's interferon-mediated immune response to CoV infection [2-4,13,15-16], so nsp13 inhibition has the potential to limit this effect on immune evasion, in addition to direct anti-replication activity. |
References
1. Corona A, Madia VN, De Santis R, Manelfi C, Emmolo R, Ialongo D, Patacchini E, Messore A, Amatore D, Faggioni G et al.. (2023) Diketo acid inhibitors of nsp13 of SARS-CoV-2 block viral replication. Antiviral Res, 217: 105697. [PMID:37562607]
2. Feng K, Min YQ, Sun X, Deng F, Li P, Wang H, Ning YJ. (2021) Interactome profiling reveals interaction of SARS-CoV-2 NSP13 with host factor STAT1 to suppress interferon signaling. J Mol Cell Biol, 13 (10): 760-762. [PMID:34687317]
3. Feng K, Zhang HJ, Min YQ, Zhou M, Deng F, Wang HL, Li PQ, Ning YJ. (2023) SARS-CoV-2 NSP13 interacts with host IRF3, blocking antiviral immune responses. J Med Virol, 95 (6): e28881. [PMID:37314155]
4. Fung SY, Siu KL, Lin H, Chan CP, Yeung ML, Jin DY. (2022) SARS-CoV-2 NSP13 helicase suppresses interferon signaling by perturbing JAK1 phosphorylation of STAT1. Cell Biosci, 12 (1): 36. [PMID:35317858]
5. Gervasoni S, Vistoli G, Talarico C, Manelfi C, Beccari AR, Studer G, Tauriello G, Waterhouse AM, Schwede T, Pedretti A. (2020) A Comprehensive Mapping of the Druggable Cavities within the SARS-CoV-2 Therapeutically Relevant Proteins by Combining Pocket and Docking Searches as Implemented in Pockets 2.0. Int J Mol Sci, 21 (14): 5152. [PMID:32708196]
6. Inniss NL, Rzhetskaya M, Ling-Hu T, Lorenzo-Redondo R, Bachta KE, Satchell KJF, Hultquist JF. (2024) Activity and inhibition of the SARS-CoV-2 Omicron nsp13 R392C variant using RNA duplex unwinding assays. SLAS Discov, 29 (3): 100145. [PMID:38301954]
7. Keum YS, Jeong YJ. (2012) Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem Pharmacol, 84 (10): 1351-8. [PMID:22935448]
8. Lee C, Lee JM, Lee NR, Jin BS, Jang KJ, Kim DE, Jeong YJ, Chong Y. (2009) Aryl diketoacids (ADK) selectively inhibit duplex DNA-unwinding activity of SARS coronavirus NTPase/helicase. Bioorg Med Chem Lett, 19 (6): 1636-8. [PMID:19233643]
9. Marx SK, Mickolajczyk KJ, Craig JM, Thomas CA, Pfeffer AM, Abell SJ, Carrasco JD, Franzi MC, Huang JR, Kim HC et al.. (2023) Observing inhibition of the SARS-CoV-2 helicase at single-nucleotide resolution. Nucleic Acids Res, 51 (17): 9266-9278. [PMID:37560916]
10. Mehyar N. (2023) Coronaviruses SARS-CoV, MERS-CoV, and SARS-CoV-2 helicase inhibitors: a systematic review of invitro studies. J Virus Erad, 9 (2): 100327. [PMID:37363132]
11. Newman JA, Douangamath A, Yadzani S, Yosaatmadja Y, Aimon A, Brandão-Neto J, Dunnett L, Gorrie-Stone T, Skyner R, Fearon D et al.. (2021) Structure, mechanism and crystallographic fragment screening of the SARS-CoV-2 NSP13 helicase. Nat Commun, 12 (1): 4848. [PMID:34381037]
12. Ramsey JR, Shelton PMM, Heiss TK, Olinares PDB, Vostal LE, Soileau H, Grasso M, Casebeer SW, Adaniya S, Miller M et al.. (2024) Using a Function-First "Scout Fragment"-Based Approach to Develop Allosteric Covalent Inhibitors of Conformationally Dynamic Helicase Mechanoenzymes. J Am Chem Soc, 146 (1): 62-67. DOI: 10.1101/2023.09.25.559391 [PMID:38134034]
13. Rashid F, Xie Z, Suleman M, Shah A, Khan S, Luo S. (2022) Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front Immunol, 13: 940756. [PMID:36003396]
14. Shiryaev V, Klimochkin Y. (2024) Computer-aided Design of Wide-spectrum Coronavirus Helicase NSP13 Cage Inhibitors: A Molecular Modelling Approach. Curr Comput Aided Drug Des, 20 (7): 1027-1041. [PMID:37921184]
15. Sui C, Xiao T, Zhang S, Zeng H, Zheng Y, Liu B, Xu G, Gao C, Zhang Z. (2022) SARS-CoV-2 NSP13 Inhibits Type I IFN Production by Degradation of TBK1 via p62-Dependent Selective Autophagy. J Immunol, 208 (3): 753-761. [PMID:34996837]
16. Vazquez C, Swanson SE, Negatu SG, Dittmar M, Miller J, Ramage HR, Cherry S, Jurado KA. (2021) SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PLoS One, 16 (6): e0253089. [PMID:34166398]
17. White MA, Lin W, Cheng X. (2020) Discovery of COVID-19 Inhibitors Targeting the SARS-CoV-2 Nsp13 Helicase. J Phys Chem Lett, 11 (21): 9144-9151. [PMID:33052685]
18. Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J et al.. (2020) Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe, 27 (3): 325-328. [PMID:32035028]
19. Yu MS, Lee J, Lee JM, Kim Y, Chin YW, Jee JG, Keum YS, Jeong YJ. (2012) Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg Med Chem Lett, 22 (12): 4049-54. [PMID:22578462]
20. Zaher NH, Mostafa MI, Altaher AY. (2020) Design, synthesis and molecular docking of novel triazole derivatives as potential CoV helicase inhibitors. Acta Pharm, 70 (2): 145-159. [PMID:31955138]