GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
Contents:
Previous and Unofficial Names  |
| non-structural protein 3 | NS3 | nsp3 | PL-PRO |
Database Links  |
|
| UniProtKB | P0DTC1 (SARS-CoV-2) |
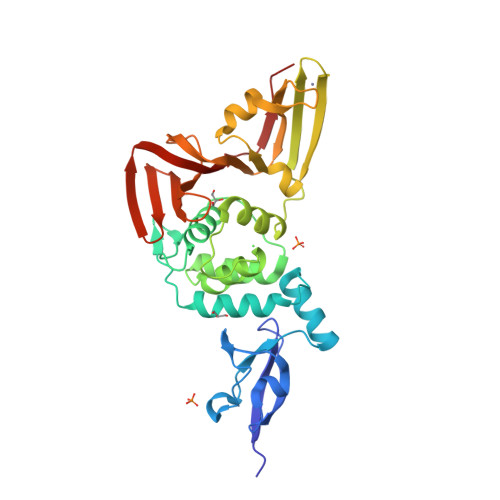
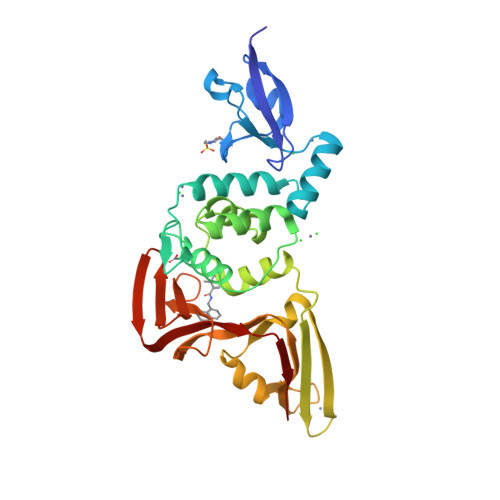
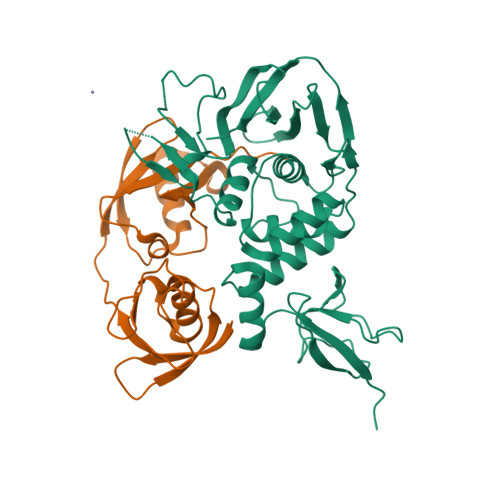
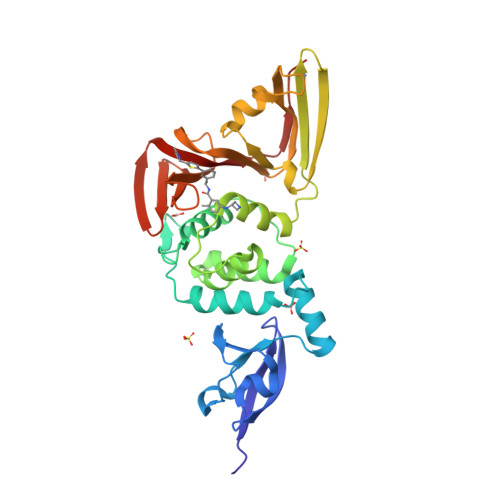
Selected 3D Structures  |
|||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
|
|
||||||||||||
| EC number (SARS-CoV-2) |
| 3.4.22.46 |
Download all structure-activity data for this target as a CSV file 
| Inhibitors | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific inhibitor tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulators | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||
| General Comments |
|
SARS-CoV-2 papain-like protease (PLpro) is translated as part of the virus' replicase polyprotein 1a/1ab, and is part of non-structural protein 3 (nsp3). It does not have a gene of its own, but we have curated it as a discrete entity to help make evolving biological and pharmacological information more accessible. PLpro is a 1945 amino acid protein, comprising aa 819-2763 of the replicase polyprotein (PRO_0000449637 in UniProt, RefSeq protein YP_009742610). Both Mpro (3CLpro; the other essential SARS-CoV-2 protease) and PLpro are autocatalytically processed from the replicase polyprotein. Coronavirus PLpros belong to the peptidase clan CA (family C16). The active site contains a classic catalytic Cys-His-Asp consensus sequence. PLpro is a multi-functional protein that catalyses cleavages within the N-terminus of the replicase polyprotein, at the consensus cleavage motif LXGG|(N/K/X) which is present between nsp1/2, nsp2/3, and nsp3/4. Based on similarity with SARS-CoV PLpro, SARS-CoV-2 PLpro likely has deubiquitinating and deISGylating activities [2,8,11], participates in assembly of new viral particles, and contributes to virus-mediated inhibition of the host innate immune type I interferon response [5,20]. Existing evidence of the druggability of SARS-CoV PLpro [12] has led to investigation of the SARS-CoV-2 homologue as a molecular target for COVID-19 therapy [6]. To date (mid-2022) PLpro appears to be conserved in SARS-CoV-2 variants; of the identified PLpro mutations, the majority are located away from the currently-targeted drug-binding site. PLpro contains a viral macrodomain that has been identified as an additional druggable region [14]. Viral macrodomains are present primarily in corona, alpha, rubi, and herpes viruses, and they act to circumvent the host innate immune defence mechanism. Small molecule inhibitors that bind to this domain would be expected to compromise the pathogenicity of SARS-CoV-2 (and potentially other coronaviruses). Inhibitors: Several hepatitis C virus (HCV) inhibitor drugs (simeprevir, vaniprevir, paritaprevir, and grazoprevir) have been reported to inhibit PLpro and SARS-CoV-2 infection in vitro [1]. These drugs were found to inhibit SARS-CoV-2 replication synergistically with the viral polymerase inhibitor remdesivir. The search for novel, selective PLpro inhibitors has revealed compounds such as GRL-0617 and compound 19 [PMID: 33979649]. Many more covalent and noncovalent PLpro inhibitors are included in Tan et al.'s 2022 'Progress and Challenges' article [20]. |
References
1. Bafna K, White K, Harish B, Rosales R, Ramelot TA, Acton TB, Moreno E, Kehrer T, Miorin L, Royer CA et al.. (2021) Hepatitis C virus drugs that inhibit SARS-CoV-2 papain-like protease synergize with remdesivir to suppress viral replication in cell culture. Cell Rep, 35 (7): 109133. [PMID:33984267]
2. Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. (2005) The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J Virol, 79 (24): 15189-98. [PMID:16306590]
3. Cockram PE, Walters BT, Lictao A, Shanahan F, Wertz IE, Foster SA, Rudolph J. (2024) Allosteric Inhibitors of the SARS-COV-2 Papain-like Protease Domain Induce Proteasomal Degradation of Its Parent Protein NSP3. ACS Chem Biol, 19 (1): 22-36. [PMID:38150587]
4. Dang F, Bai L, Dong J, Hu X, Wang J, Paulo JA, Xiong Y, Liang X, Sun Y, Chen Y et al.. (2023) USP2 inhibition prevents infection with ACE2-dependent coronaviruses in vitro and is protective against SARS-CoV-2 in mice. Sci Transl Med, 15 (725): eadh7668. [PMID:38055802]
5. Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC et al.. (2007) Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem, 282 (44): 32208-21. [PMID:17761676]
6. Freitas BT, Durie IA, Murray J, Longo JE, Miller HC, Crich D, Hogan RJ, Tripp RA, Pegan SD. (2020) Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect Dis, 6 (8): 2099-2109. [PMID:32428392]
7. Garnsey MR, Robinson MC, Nguyen LT, Cardin R, Tillotson J, Mashalidis E, Yu A, Aschenbrenner L, Balesano A, Behzadi A et al.. (2024) Discovery of SARS-CoV-2 papain-like protease (PLpro) inhibitors with efficacy in a murine infection model. bioRxiv, Preprint. DOI: 10.1101/2024.01.26.577395
8. Lindner HA, Lytvyn V, Qi H, Lachance P, Ziomek E, Ménard R. (2007) Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch Biochem Biophys, 466 (1): 8-14. [PMID:17692280]
9. M Bader S, Calleja DJ, Devine SM, Kuchel NW, Lu BGC, Wu X, Birkinshaw RW, Bhandari R, Loi K, Volpe R et al.. (2025) A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID. Nat Commun, 16 (1): 2900. [PMID:40180914]
10. Osipiuk J, Azizi SA, Dvorkin S, Endres M, Jedrzejczak R, Jones KA, Kang S, Kathayat RS, Kim Y, Lisnyak VG et al.. (2021) Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nat Commun, 12 (1): 743. [PMID:33531496]
11. Osipiuk J, Wydorski PM, Lanham BT, Tesar C, Endres M, Engle E, Jedrzejczak R, Mullapudi V, Michalska K, Fidelis K et al.. (2022) Dual domain recognition determines SARS-CoV-2 PLpro selectivity for human ISG15 and K48-linked di-ubiquitin. bioRxiv, Preprint. DOI: 10.1101/2021.09.15.460543 [PMID:35547846]
12. Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME et al.. (2008) A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc Natl Acad Sci USA, 105 (42): 16119-24. [PMID:18852458]
13. Sanders BC, Pokhrel S, Labbe AD, Mathews II, Cooper CJ, Davidson RB, Phillips G, Weiss KL, Zhang Q, O'Neill H et al.. (2023) Potent and selective covalent inhibition of the papain-like protease from SARS-CoV-2. Nat Commun, 14 (1): 1733. [PMID:36977673]
14. Schuller M, Correy GJ, Gahbauer S, Fearon D, Wu T, Diaz RE, Young ID, Martins LC, Smith DH, Schulze-Gahmen U et al.. (2021) Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Science Advances, 7 (16): eabf8711 [Epub ahead of print]. DOI: 10.1126/sciadv.abf8711
15. Shan H, Liu J, Shen J, Dai J, Xu G, Lu K, Han C, Wang Y, Xu X, Tong Y et al.. (2021) Development of potent and selective inhibitors targeting the papain-like protease of SARS-CoV-2. Cell Chem Biol, 28 (6): 855-865.e9. [PMID:33979649]
16. Shen Z, Ratia K, Cooper L, Kong D, Lee H, Kwon Y, Li Y, Alqarni S, Huang F, Dubrovskyi O et al.. (2021) Potent, Novel SARS-CoV-2 PLpro Inhibitors Block Viral Replication in Monkey and Human Cell Cultures. bioRxiv,. [PMID:33594371]
17. Shen Z, Ratia K, Cooper L, Kong D, Lee H, Kwon Y, Li Y, Alqarni S, Huang F, Dubrovskyi O et al.. (2022) Design of SARS-CoV-2 PLpro Inhibitors for COVID-19 Antiviral Therapy Leveraging Binding Cooperativity. J Med Chem, 65 (4): 2940-2955. [PMID:34665619]
18. Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G et al.. (2020) Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature, 587 (7835): 657-662. [PMID:32726803]
19. Tan B, Zhang X, Ansari A, Jadhav P, Tan H, Li K, Chopra A, Ford A, Chi X, Ruiz FX et al.. (2024) Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model. Science, 383 (6690): 1434-1440. [PMID:38547259]
20. Tan H, Hu Y, Jadhav P, Tan B, Wang J. (2022) Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease. J Med Chem, 65 (11): 7561-7580. [PMID:35620927]
21. Węglarz-Tomczak E, Tomczak JM, Talma M, Brul S. (2020) Ebselen as a highly active inhibitor of PLProCoV2. bioRxiv, Preprint. DOI: 10.1101/2020.05.17.100768