GtoPdb is requesting financial support from commercial users. Please see our sustainability page for more information.
|
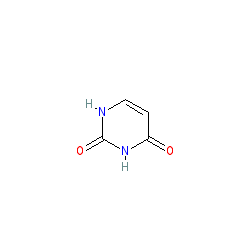
Compound class:
Metabolite
|
|
|||||||||||||||||||||||||||||||||||
| References |
|
1. Yamamoto S, Inoue K, Murata T, Kamigaso S, Yasujima T, Maeda JY, Yoshida Y, Ohta KY, Yuasa H. (2010)
Identification and functional characterization of the first nucleobase transporter in mammals: implication in the species difference in the intestinal absorption mechanism of nucleobases and their analogs between higher primates and other mammals. J Biol Chem, 285 (9): 6522-31. [PMID:20042597] |