|
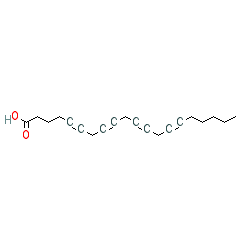
Abbreviated name: ETYA
Compound class:
Metabolite
Ligand Activity Visualisation ChartsThese are box plot that provide a unique visualisation, summarising all the activity data for a ligand taken from ChEMBL and GtoPdb across multiple targets and species. Click on a plot to see the median, interquartile range, low and high data points. A value of zero indicates that no data are available. A separate chart is created for each target, and where possible the algorithm tries to merge ChEMBL and GtoPdb targets by matching them on name and UniProt accession, for each available species. However, please note that inconsistency in naming of targets may lead to data for the same target being reported across multiple charts. ✖ |
|
|||||||||||||||||||||||||||||||||||
| References |
|
1. Keller H, Devchand PR, Perroud M, Wahli W. (1997)
PPAR alpha structure-function relationships derived from species-specific differences in responsiveness to hypolipidemic agents. Biol Chem, 378 (7): 651-5. [PMID:9278144] |
|
2. Keller H, Dreyer C, Medin J, Mahfoudi A, Ozato K, Wahli W. (1993)
Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc Natl Acad Sci USA, 90 (6): 2160-4. [PMID:8384714] |