Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Natural/Endogenous Ligands
- Agonists
- Antagonists
- Channel Blockers
- Allosteric Modulators
- Immuno Process Associations
- Clinically-Relevant Mutations and Pathophysiology
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
|||||||
| Species | TM | P Loops | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 3 | 1 | 1484 | 12p13.1 | GRIN2B | glutamate ionotropic receptor NMDA type subunit 2B | |
| Mouse | 3 | 1 | 1482 | 6 66.38 cM | Grin2b | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | |
| Rat | 3 | 1 | 1482 | 4q43 | Grin2b | glutamate ionotropic receptor NMDA type subunit 2B | |
Previous and Unofficial Names  |
| NMDAR2B | GluRepsilon2 | glutamate receptor, ionotropic, N-methyl D-aspartate 2B | glutamate receptor, ionotropic, NMDA2B (epsilon 2) | glutamate receptor |
Database Links  |
|
| Alphafold | Q13224 (Hs), Q01097 (Mm), Q00960 (Rn) |
| ChEMBL Target | CHEMBL1904 (Hs), CHEMBL3442 (Mm), CHEMBL311 (Rn) |
| DrugBank Target | Q13224 (Hs), Q13224 (Hs) |
| Ensembl Gene | ENSG00000273079 (Hs), ENSMUSG00000030209 (Mm), ENSRNOG00000008766 (Rn) |
| Entrez Gene | 2904 (Hs), 14812 (Mm), 24410 (Rn) |
| Human Protein Atlas | ENSG00000273079 (Hs) |
| KEGG Gene | hsa:2904 (Hs), mmu:14812 (Mm), rno:24410 (Rn) |
| OMIM | 138252 (Hs) |
| Orphanet | ORPHA266126 (Hs) |
| Pharos | Q13224 (Hs) |
| RefSeq Nucleotide | NM_000834 (Hs), NM_008171 (Mm), NM_012574 (Rn) |
| RefSeq Protein | NP_000825 (Hs), NP_032197 (Mm), NP_036706 (Rn) |
| UniProtKB | Q13224 (Hs), Q01097 (Mm), Q00960 (Rn) |
| Wikipedia | GRIN2B (Hs) |
Selected 3D Structures  |
|||||||||||||
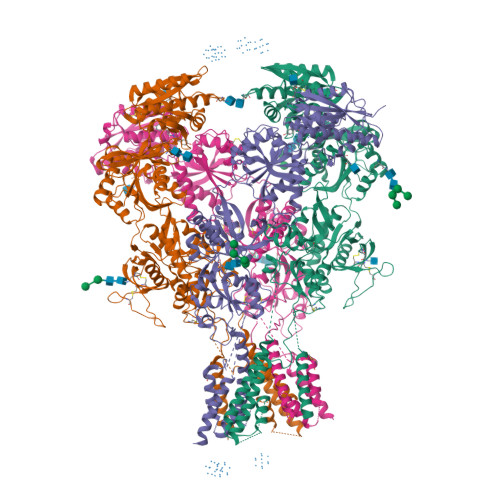
|
|
||||||||||||
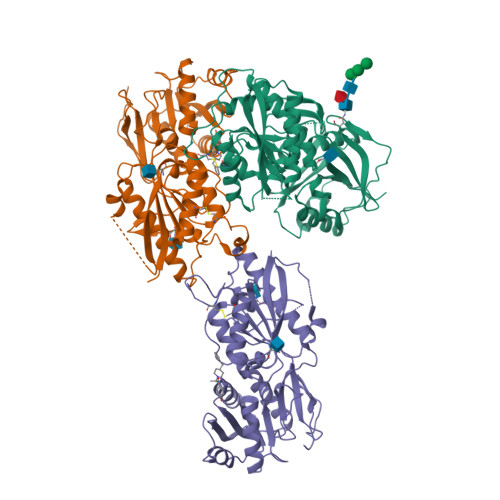
|
|
||||||||||||
Natural/Endogenous Ligands  |
| D-aspartic acid |
| D-serine |
| glycine |
| L-aspartic acid |
| L-glutamic acid |
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Channel Blockers | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Allosteric Modulators | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific allosteric modulator tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Immuno Process Associations | ||
|
Clinically-Relevant Mutations and Pathophysiology 
|
||||||||||||||
|
||||||||||||||
References
1. Štefková-Mazochová K, Danda H, Dehaen W, Jurásek B, Šíchová K, Pinterová-Leca N, Mazoch V, Krausová BH, Kysilov B, Smejkalová T et al.. (2022) Pharmacokinetic, pharmacodynamic, and behavioural studies of deschloroketamine in Wistar rats. Br J Pharmacol, 179 (1): 65-83. [PMID:34519023]
2. Acker TM, Yuan H, Hansen KB, Vance KM, Ogden KK, Jensen HS, Burger PB, Mullasseril P, Snyder JP, Liotta DC et al.. (2011) Mechanism for noncompetitive inhibition by novel GluN2C/D N-methyl-D-aspartate receptor subunit-selective modulators. Mol Pharmacol, 80 (5): 782-95. [PMID:21807990]
3. Anderson DR, Volkmann RA, Menniti FS. (2019) N-alkylaryl-5-oxyaryl-octahydro-cyclopenta[c]pyrrole negative allosteric modulators of NR2B. Patent number: US10239835B2. Assignee: Cadent Therapeutics Inc. Priority date: 26/09/2014. Publication date: 26/03/2019.
4. Auberson YP, Allgeier H, Bischoff S, Lingenhoehl K, Moretti R, Schmutz M. (2002) 5-Phosphonomethylquinoxalinediones as competitive NMDA receptor antagonists with a preference for the human 1A/2A, rather than 1A/2B receptor composition. Bioorg Med Chem Lett, 12 (7): 1099-102. [PMID:11909726]
5. Feng B, Tse HW, Skifter DA, Morley R, Jane DE, Monaghan DT. (2004) Structure-activity analysis of a novel NR2C/NR2D-preferring NMDA receptor antagonist: 1-(phenanthrene-2-carbonyl) piperazine-2,3-dicarboxylic acid. Br J Pharmacol, 141 (3): 508-16. [PMID:14718249]
6. Frizelle PA, Chen PE, Wyllie DJ. (2006) Equilibrium constants for (R)-[(S)-1-(4-bromo-phenyl)-ethylamino]-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-5-yl)-methyl]-phosphonic acid (NVP-AAM077) acting at recombinant NR1/NR2A and NR1/NR2B N-methyl-D-aspartate receptors: Implications for studies of synaptic transmission. Mol Pharmacol, 70 (3): 1022-32. [PMID:16778008]
7. Hill MD, Blanco MJ, Salituro FG, Bai Z, Beckley JT, Ackley MA, Dai J, Doherty JJ, Harrison BL, Hoffmann EC et al.. (2022) SAGE-718: A First-in-Class N-Methyl-d-Aspartate Receptor Positive Allosteric Modulator for the Potential Treatment of Cognitive Impairment. J Med Chem, 65 (13): 9063-9075. [PMID:35785990]
8. Karakas E, Furukawa H. (2014) Crystal structure of a heterotetrameric NMDA receptor ion channel. Science, 344 (6187): 992-7. [PMID:24876489]
9. Karakas E, Simorowski N, Furukawa H. (2011) Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature, 475 (7355): 249-53. [PMID:21677647]
10. Morley RM, Tse HW, Feng B, Miller JC, Monaghan DT, Jane DE. (2005) Synthesis and pharmacology of N1-substituted piperazine-2,3-dicarboxylic acid derivatives acting as NMDA receptor antagonists. J Med Chem, 48 (7): 2627-37. [PMID:15801853]
11. Neyton J, Paoletti P. (2006) Relating NMDA receptor function to receptor subunit composition: limitations of the pharmacological approach. J Neurosci, 26 (5): 1331-3. [PMID:16452656]