Contents:
- Gene and Protein Information
- Previous and Unofficial Names
- Database Links
- Selected 3D Structures
- Associated Proteins
- Natural/Endogenous Ligands
- Rank order lists
- Agonists
- Antagonists
- Transduction Mechanisms
- Tissue Distribution
- Expression Datasets
- Functional Assays
- Physiological Functions
- Physiological Consequences of Altering Gene Expression
- Phenotypes, Alleles and Disease Models
- Biologically Significant Variants
- General Comments
- References
- Contributors
- How to cite this page
Gene and Protein Information  |
||||||
| class A G protein-coupled receptor | ||||||
| Species | TM | AA | Chromosomal Location | Gene Symbol | Gene Name | Reference |
| Human | 7 | 418 | 20q13.33 | NTSR1 | neurotensin receptor 1 | 43-44,70 |
| Mouse | 7 | 424 | 2 103.04 cM | Ntsr1 | neurotensin receptor 1 | 43 |
| Rat | 7 | 424 | 3q43 | Ntsr1 | neurotensin receptor 1 | 68 |
Previous and Unofficial Names  |
|
| NTR1 | NTRH [8] | NTSR1 | NTR | neurotensin receptor type 1 | Levocabastin-insensitive neurotensin receptor | NT-1R | neurotensin receptor 1 (high affinity) | |
Database Links  |
|
| Specialist databases | |
| GPCRdb | ntr1_human (Hs), ntr1_mouse (Mm), ntr1_rat (Rn) |
| Other databases | |
| Alphafold | P30989 (Hs), O88319 (Mm), P20789 (Rn) |
| ChEMBL Target | CHEMBL4123 (Hs), CHEMBL3570 (Mm), CHEMBL3027 (Rn) |
| Ensembl Gene | ENSG00000101188 (Hs), ENSMUSG00000027568 (Mm), ENSRNOG00000028708 (Rn) |
| Entrez Gene | 4923 (Hs), 18216 (Mm), 366274 (Rn) |
| Human Protein Atlas | ENSG00000101188 (Hs) |
| KEGG Gene | hsa:4923 (Hs), mmu:18216 (Mm), rno:366274 (Rn) |
| OMIM | 162651 (Hs) |
| Pharos | P30989 (Hs) |
| RefSeq Nucleotide | NM_002531 (Hs), NM_018766 (Mm), NM_001108967 (Rn) |
| RefSeq Protein | NP_002522 (Hs), NP_061236 (Mm), NP_001102437 (Rn) |
| UniProtKB | P30989 (Hs), O88319 (Mm), P20789 (Rn) |
| Wikipedia | NTSR1 (Hs) |
Selected 3D Structures  |
|||||||||||||
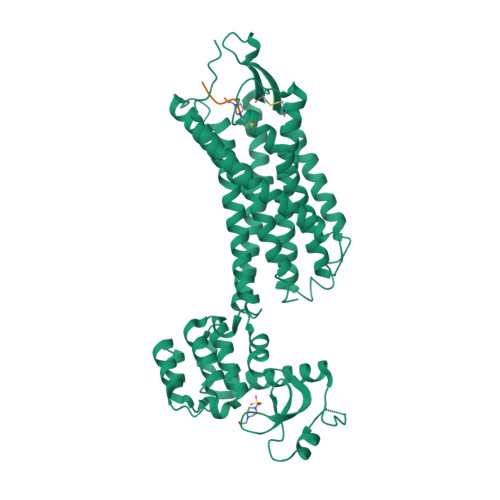
|
|
||||||||||||
Associated Proteins  |
|||||||||
|
|||||||||
Natural/Endogenous Ligands  |
| large neuromedin N {Sp: Human} , large neuromedin N {Sp: Mouse} , large neuromedin N {Sp: Rat} |
| large neurotensin {Sp: Human} |
| neuromedin N {Sp: Human} , neuromedin N {Sp: Mouse, Rat} |
| neurotensin {Sp: Human, Mouse, Rat, Bovine} |
| Comments: Neurotensin is the most potent endogenous agonist |
| Potency order of endogenous ligands (Human) |
| neurotensin (NTS, P30990) > neuromedin N [38] |
Download all structure-activity data for this target as a CSV file 
| Agonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific agonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Agonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| The sequence of neurotensin is identical for all mammalian species. Neurotensin and neuromedin N are synthesised from a common precursor [24]. Neurotensin and [125I]neurotensin bind with high affinity to NTS1 from all species [125I]-Trp11-neurotensin is specific for murine NTS1 receptors and exhibits low affinity for NTS1 receptors of human or other species [48,59]. [125I]-azidobenzoyl-neurotensin(2-13) is a photoreactive and radioactive analogue for covalent labelling of neurotensin receptors (including NTS1) from any species [15-16]. Xenopsin and contulakin-G are natural analogues of neurotensin from Xenopus laevis [5] and Conus geographus resepctively [20]. [α-bodipy]neurotensin(2-13) is a fluorescent synthetic analogue of neurotensin [7]. KH28, ABS212 and ABS-201 are able to cross the blood-brain barrier. ABS-201 and ABS212 are stable bioavailable NT analogues. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonists | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Key to terms and symbols | View all chemical structures | Click column headers to sort | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| View species-specific antagonist tables | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Antagonist Comments | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SR142948A binds to both NTS1 and NTS2 with similar nanomolar affinities. SR48692 has a 10-30 times higher affinity for NTS1 than for NTS2 [35,42]. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Primary Transduction Mechanisms 
|
|
| Transducer | Effector/Response |
| Gq/G11 family | Phospholipase C stimulation |
| References: 2-3,32,39,53,67,71 | |
Secondary Transduction Mechanisms  |
|
| Transducer | Effector/Response |
|
Gs family Gi/Go family |
Adenylyl cyclase stimulation Adenylyl cyclase inhibition Guanylate cyclase stimulation Other - See Comments |
| Comments: Binding of neurotensin to the human NTS1 receptor indicates activation of MAP kinases and stimulates transcription of Krox-24, c-fos and Elk-1 [56,58]. | |
| References: 4,14,32-33,53,73 | |
Tissue Distribution 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Expression Datasets  |
|
|
Functional Assays 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Physiological Functions 
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
||||||||
|
Physiological Consequences of Altering Gene Expression 
|
||||||||||
|
||||||||||
|
||||||||||
|
||||||||||
|
Phenotypes, Alleles and Disease Models 
|
Mouse data from MGI | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Biologically Significant Variants 
|
||||||||
|
| General Comments |
| A special issue of the journal "peptides" entitled "Neurotensin: Roles and Mechanisms" has recently been published [26]. |
References
1. Alexander MJ, Leeman SE. (1998) Widespread expression in adult rat forebrain of mRNA encoding high-affinity neurotensin receptor. J Comp Neurol, 402 (4): 475-500. [PMID:9862322]
2. Amar S, Kitabgi P, Vincent JP. (1986) Activation of phosphatidylinositol turnover by neurotensin receptors in the human colonic adenocarcinoma cell line HT29. FEBS Lett, 201 (1): 31-6. [PMID:3011505]
3. Amar S, Kitabgi P, Vincent JP. (1987) Stimulation of inositol phosphate production by neurotensin in neuroblastoma N1E115 cells: implication of GTP-binding proteins and relationship with the cyclic GMP response. J Neurochem, 49 (4): 999-1006. [PMID:3040912]
4. Amar S, Mazella J, Checler F, Kitabgi P, Vincent JP. (1985) Regulation of cyclic GMP levels by neurotensin in neuroblastoma clone N1E115. Biochem Biophys Res Commun, 129 (1): 117-25. [PMID:2988544]
5. Araki K, Tachibana S, Uchiyama M, Nakajima T, Yasuhara T. (1975) Isolation and structure of a new active peptide xenopsin on rat stomach strip and some biogenic amines in the skin of Xenopus laevis. Chem Pharm Bull (Tokyo), 23: 3132-3140. [PMID:1218451]
6. Azriel Y, Burcher E. (2001) Characterization and autoradiographic localization of neurotensin binding sites in human sigmoid colon. J Pharmacol Exp Ther, 297 (3): 1074-81. [PMID:11356931]
7. Beaudet A, Nouel D, Stroh T, Vandenbulcke F, Dal-Farra C, Vincent JP. (1998) Fluorescent ligands for studying neuropeptide receptors by confocal microscopy. Braz J Med Biol Res, 31 (11): 1479-89. [PMID:9921286]
8. Botto JM, Guillemare E, Vincent JP, Mazella J. (1997) Effects of SR 48692 on neurotensin-induced calcium-activated chloride currents in the Xenopus oocyte expression system: agonist-like activity on the levocabastine-sensitive neurotensin receptor and absence of antagonist effect on the levocabastine insensitive neurotensin receptor. Neurosci Lett, 223 (3): 193-6. [PMID:9080465]
9. Botto JM, Vincent JP, Mazella J. (1997) Existence of two translation initiation sites leading to the expression of two proteins from the rat high-affinity neurotensin-receptor cDNA: possible regulation by the 5' end non-coding region. Biochem J, 324 ( Pt 2): 389-93. [PMID:9182695]
10. Boudin H, Lazaroff B, Bachelet CM, Pélaprat D, Rostène W, Beaudet A. (2000) Immunologic differentiation of two high-affinity neurotensin receptor isoforms in the developing rat brain. J Comp Neurol, 425 (1): 45-57. [PMID:10940941]
11. Boudin H, Pélaprat D, Rostène W, Beaudet A. (1996) Cellular distribution of neurotensin receptors in rat brain: immunohistochemical study using an antipeptide antibody against the cloned high affinity receptor. J Comp Neurol, 373 (1): 76-89. [PMID:8876464]
12. Boudin H, Pélaprat D, Rostène W, Pickel VM, Beaudet A. (1998) Correlative ultrastructural distribution of neurotensin receptor proteins and binding sites in the rat substantia nigra. J Neurosci, 18 (20): 8473-84. [PMID:9763490]
13. Boules M, Shaw A, Liang Y, Barbut D, Richelson E. (2009) NT69L, a novel analgesic, shows synergy with morphine. Brain Res, 1294: 22-8. [PMID:19651107]
14. Bozou JC, Amar S, Vincent JP, Kitabgi P. (1986) Neurotensin-mediated inhibition of cyclic AMP formation in neuroblastoma N1E115 cells: involvement of the inhibitory GTP-binding component of adenylate cyclase. Mol Pharmacol, 29 (5): 489-96. [PMID:3010077]
15. Chabry J, Gaudriault G, Vincent JP, Mazella J. (1993) Implication of various forms of neurotensin receptors in the mechanism of internalization of neurotensin in cerebral neurons. J Biol Chem, 268 (23): 17138-44. [PMID:8394329]
16. Chabry J, Labbé-Jullié C, Gully D, Kitabgi P, Vincent JP, Mazella J. (1994) Stable expression of the cloned rat brain neurotensin receptor into fibroblasts: binding properties, photoaffinity labeling, transduction mechanisms, and internalization. J Neurochem, 63: 19-27. [PMID:8207427]
17. Choi KE, Hall CL, Sun JM, Wei L, Mohamad O, Dix TA, Yu SP. (2012) A novel stroke therapy of pharmacologically induced hypothermia after focal cerebral ischemia in mice. FASEB J, 26 (7): 2799-810. [PMID:22459147]
18. Choi SY, Chae HD, Park TJ, Ha H, Kim KT. (1999) Characterization of high affinity neurotensin receptor NTR1 in HL-60 cells and its down regulation during granulocytic differentiation. Br J Pharmacol, 126 (4): 1050-6. [PMID:10193787]
19. Coppola T, Béraud-Dufour S, Antoine A, Vincent JP, Mazella J. (2008) Neurotensin protects pancreatic beta cells from apoptosis. Int J Biochem Cell Biol, 40 (10): 2296-302. [PMID:18456542]
20. Craig AG, Norberg T, Griffin D, Hoeger C, Akhtar M, Schmidt K, Low W, Dykert J, Richelson E, Navarro V et al.. (1999) Contulakin-G, an O-glycosylated invertebrate neurotensin. J Biol Chem, 274 (20): 13752-9. [PMID:10318778]
21. Croci T, Aureggi G, Guagnini F, Manara L, Gully D, Fur GL, Maffrand JP, Mukenge S, Ferla G, Ferrara P et al.. (1999) In vitro functional evidence of different neurotensin-receptors modulating the motor response of human colonic muscle strips. Br J Pharmacol, 127 (8): 1922-8. [PMID:10482925]
22. Cusack B, Jansen K, McCormick DJ, Chou T, Pang Y, Richelson E. (2000) A single amino acid of the human and rat neurotensin receptors (subtype 1) determining the pharmacological profile of a species-selective neurotensin agonist. Biochem Pharmacol, 60 (6): 793-801. [PMID:10930533]
23. Cáceda R, Kinkead B, Owens MJ, Nemeroff CB. (2005) Virally mediated increased neurotensin 1 receptor in the nucleus accumbens decreases behavioral effects of mesolimbic system activation. J Neurosci, 25 (50): 11748-56. [PMID:16354933]
24. Dobner PR, Barber DL, Villa-Komaroff L, McKiernan C. (1987) Cloning and sequence analysis of cDNA for the canine neurotensin/neuromedin N precursor. Proc Natl Acad Sci USA, 84 (10): 3516-20. [PMID:3472221]
25. Dubuc I, Sarret P, Labbé-Jullié C, Botto JM, Honoré E, Bourdel E, Martinez J, Costentin J, Vincent JP, Kitabgi P et al.. (1999) Identification of the receptor subtype involved in the analgesic effect of neurotensin. J Neurosci, 19 (1): 503-10. [PMID:9870978]
26. edCarraway RE, Pothoulakis C, Leeman S. (2006) Neurotensin: Roles and Mechanisms. Peptides, 27: 2361-2534.
27. Elde R, Schalling M, Ceccatelli S, Nakanishi S, Hökfelt T. (1990) Localization of neuropeptide receptor mRNA in rat brain: initial observations using probes for neurotensin and substance P receptors. Neurosci Lett, 120 (1): 134-8. [PMID:1705671]
28. Fantegrossi WE, Ko MC, Woods JH, Richelson E. (2005) Antinociceptive, hypothermic, hypotensive, and reinforcing effects of a novel neurotensin receptor agonist, NT69L, in rhesus monkeys. Pharmacol Biochem Behav, 80 (2): 341-9. [PMID:15680187]
29. Fassio A, Evans G, Grisshammer R, Bolam JP, Mimmack M, Emson PC. (2000) Distribution of the neurotensin receptor NTS1 in the rat CNS studied using an amino-terminal directed antibody. Neuropharmacology, 39 (8): 1430-42. [PMID:10818259]
30. Friry C, Feliciangeli S, Richard F, Kitabgi P, Rovere C. (2002) Production of recombinant large proneurotensin/neuromedin N-derived peptides and characterization of their binding and biological activity. Biochem Biophys Res Commun, 290 (4): 1161-8. [PMID:11811984]
31. Gailly P. (1998) Ca2+ entry in CHO cells, after Ca2+ stores depletion, is mediated by arachidonic acid. Cell Calcium, 24 (4): 293-304. [PMID:9883283]
32. Gailly P, Najimi M, Hermans E. (2000) Evidence for the dual coupling of the rat neurotensin receptor with pertussis toxin-sensitive and insensitive G-proteins. FEBS Lett, 483 (2-3): 109-13. [PMID:11042263]
33. Gilbert JA, Richelson E. (1984) Neurotensin stimulates formation of cyclic GMP in murine neuroblastoma clone N1E-115. Eur J Pharmacol, 99 (2-3): 245-6. [PMID:6329783]
34. Guillemette A, Dansereau MA, Beaudet N, Richelson E, Sarret P. (2012) Intrathecal administration of NTS1 agonists reverses nociceptive behaviors in a rat model of neuropathic pain. Eur J Pain, 16 (4): 473-84. [PMID:22396077]
35. Gully D, Canton M, Boigegrain R, Jeanjean F, Molimard JC, Poncelet M, Gueudet C, Heaulme M, Leyris R, Brouard A et al.. (1993) Biochemical and pharmacological profile of a potent and selective nonpeptide antagonist of the neurotensin receptor. Proc Natl Acad Sci USA, 90 (1): 65-9. [PMID:8380498]
36. Gully D, Labeeuw B, Boigegrain R, Oury-Donat F, Bachy A, Poncelet M, Steinberg R, Suaud-Chagny MF, Santucci V, Vita N et al.. (1997) Biochemical and pharmacological activities of SR 142948A, a new potent neurotensin receptor antagonist. J Pharmacol Exp Ther, 280 (2): 802-12. [PMID:9023294]
37. Hadden MK, Orwig KS, Kokko KP, Mazella J, Dix TA. (2005) Design, synthesis, and evaluation of the antipsychotic potential of orally bioavailable neurotensin (8-13) analogues containing non-natural arginine and lysine residues. Neuropharmacology, 49 (8): 1149-59. [PMID:16095636]
38. Hermans E, Geurts M, Maloteaux JM. (1997) Agonist and antagonist modulation of [35S]-GTP gamma S binding in transfected CHO cells expressing the neurotensin receptor. Br J Pharmacol, 121 (8): 1817-23. [PMID:9283723]
39. Hermans E, Maloteaux JM, Octave JN. (1992) Phospholipase C activation by neurotensin and neuromedin N in Chinese hamster ovary cells expressing the rat neurotensin receptor. Brain Res Mol Brain Res, 15 (3-4): 332-8. [PMID:1331689]
40. Hughes FM, Shaner BE, May LA, Zotian L, Brower JO, Woods RJ, Cash M, Morrow D, Massa F, Mazella J et al.. (2010) Identification and functional characterization of a stable, centrally active derivative of the neurotensin (8-13) fragment as a potential first-in-class analgesic. J Med Chem, 53 (12): 4623-32. [PMID:20481538]
41. Labbé-Jullié C, Barroso S, Nicolas-Etève D, Reversat JL, Botto JM, Mazella J, Bernassau JM, Kitabgi P. (1998) Mutagenesis and modeling of the neurotensin receptor NTR1. Identification of residues that are critical for binding SR 48692, a nonpeptide neurotensin antagonist. J Biol Chem, 273 (26): 16351-7. [PMID:9632698]
42. Labbé-Jullié C, Botto JM, Mas MV, Chabry J, Mazella J, Vincent JP, Gully D, Maffrand JP, Kitabgi P. (1995) [3H]SR 48692, the first nonpeptide neurotensin antagonist radioligand: characterization of binding properties and evidence for distinct agonist and antagonist binding domains on the rat neurotensin receptor. Mol Pharmacol, 47 (5): 1050-6. [PMID:7746272]
43. Laurent P, Clerc P, Mattei MG, Forgez P, Dumont X, Ferrara P, Caput D, Rostene W. (1994) Chromosomal localization of mouse and human neurotensin receptor genes. Mamm Genome, 5: 303-306. [PMID:8075503]
44. Le F, Groshan K, Zeng XP, Richelson E. (1997) Characterization of the genomic structure, promoter region, and a tetranucleotide repeat polymorphism of the human neurotensin receptor gene. J Biol Chem, 272 (2): 1315-22. [PMID:8995438]
45. Leonetti M, Brun P, Clerget M, Steinberg R, Soubrié P, Renaud B, Suaud-Chagny MF. (2004) Specific involvement of neurotensin type 1 receptor in the neurotensin-mediated in vivo dopamine efflux using knock-out mice. J Neurochem, 89 (1): 1-6. [PMID:15030383]
46. Li H, Hua T, Wang W, Wu X, Miao C, Huang W, Xiao Y, Yang J, Bradley JL, Peberdy MA et al.. (2019) The Effects of Pharmacological Hypothermia Induced by Neurotensin Receptor Agonist ABS 201 on Outcomes of CPR. Shock, 51 (5): 667-673. [PMID:30986796]
47. Martin S, Navarro V, Vincent JP, Mazella J. (2002) Neurotensin receptor-1 and -3 complex modulates the cellular signaling of neurotensin in the HT29 cell line. Gastroenterology, 123 (4): 1135-43. [PMID:12360476]
48. Mazella J, Poustis C, Labbe C, Checler F, Kitabgi P, Granier C, van Rietschoten J, Vincent JP. (1983) Monoiodo-[Trp11]neurotensin, a highly radioactive ligand of neurotensin receptors. Preparation, biological activity, and binding properties to rat brain synaptic membranes. J Biol Chem, 258 (6): 3476-81. [PMID:6300046]
49. Mechanic JA, Sutton JE, Berson AE, Wu X, Kwan J, Schreiber R, Pang Z, Button DC. (2009) Involvement of the neurotensin receptor 1 in the behavioral effects of two neurotensin agonists, NT-2 and NT69L: lack of hypothermic, antinociceptive and antipsychotic actions in receptor knockout mice. Eur Neuropsychopharmacol, 19 (7): 466-75. [PMID:19223157]
50. Moyse E, Rostène W, Vial M, Leonard K, Mazella J, Kitabgi P, Vincent JP, Beaudet A. (1987) Distribution of neurotensin binding sites in rat brain: a light microscopic radioautographic study using monoiodo [125I]Tyr3-neurotensin. Neuroscience, 22 (2): 525-36. [PMID:3313097]
51. Mulè F, Serio R, Postorino A, Vetri T, Bonvissuto F. (1996) Antagonism by SR 48692 of mechanical responses to neurotensin in rat intestine. Br J Pharmacol, 117 (3): 488-492. [PMID:8821538]
52. Najimi M, Gailly P, Maloteaux JM, Hermans E. (2002) Distinct regions of C-terminus of the high affinity neurotensin receptor mediate the functional coupling with pertussis toxin sensitive and insensitive G-proteins. FEBS Lett, 512: 329-333. [PMID:11852105]
53. Oury-Donat F, Thurneyssen O, Gonalons N, Forgez P, Gully D, Le Fur G, Soubrie P. (1995) Characterization of the effect of SR48692 on inositol monophosphate, cyclic GMP and cyclic AMP responses linked to neurotensin receptor activation in neuronal and non-neuronal cells. Br J Pharmacol, 116 (2): 1899-905. [PMID:8528577]
54. Pereyra-Alfonso S, López Ordieres MG, del V Armanino M, de Lores Arnaiz GR. (2005) High-affinity neurotensin receptor is involved in phosphoinositide hydrolysis stimulation by carbachol in neonatal rat brain. Brain Res Dev Brain Res, 154 (2): 247-54. [PMID:15707678]
55. Pettibone DJ, Hess JF, Hey PJ, Jacobson MA, Leviten M, Lis EV, Mallorga PJ, Pascarella DM, Snyder MA, Williams JB et al.. (2002) The effects of deleting the mouse neurotensin receptor NTR1 on central and peripheral responses to neurotensin. J Pharmacol Exp Ther, 300 (1): 305-13. [PMID:11752130]
56. Poinot-Chazel C, Portier M, Bouaboula M, Vita N, Pecceu F, Gully D, Monroe JG, Maffrand JP, Le Fur G, Casellas P. (1996) Activation of mitogen-activated protein kinase couples neurotensin receptor stimulation to induction of the primary response gene Krox-24. Biochem J, 320 ( Pt 1): 145-51. [PMID:8947479]
57. Poncelet M, Souilhac J, Gueudet C, Terranova JP, Gully D, Le Fur G, Soubrié P. (1994) Effects of SR 48692, a selective non-peptide neurotensin receptor antagonist, on two dopamine-dependent behavioural responses in mice and rats. Psychopharmacology (Berl.), 116 (2): 237-41. [PMID:7862953]
58. Portier M, Combes T, Gully D, Maffrand JP, Casellas P. (1998) Neurotensin type 1 receptor-mediated activation of krox24, c-fos and Elk-1: preventing effect of the neurotensin antagonists SR 48692 and SR 142948. FEBS Lett, 432 (1-2): 88-93. [PMID:9710257]
59. Poustis C, Mazella J, Kitabgi P, Vincent JP. (1984) High-affinity neurotensin binding sites in differentiated neuroblastoma N1E115 cells. J Neurochem, 42 (4): 1094-100. [PMID:6699640]
60. Remaury A, Vita N, Gendreau S, Jung M, Arnone M, Poncelet M, Culouscou JM, Le Fur G, Soubrié P, Caput D et al.. (2002) Targeted inactivation of the neurotensin type 1 receptor reveals its role in body temperature control and feeding behavior but not in analgesia. Brain Res, 953 (1-2): 63-72. [PMID:12384239]
61. Richard F, Barroso S, Martinez J, Labbé-Jullié C, Kitabgi P. (2001) Agonism, inverse agonism, and neutral antagonism at the constitutively active human neurotensin receptor 2. Mol Pharmacol, 60 (6): 1392-8. [PMID:11723247]
62. Roussy G, Beaudry H, Lafrance M, Belleville K, Beaudet N, Wada K, Gendron L, Sarret P. (2010) Altered morphine-induced analgesia in neurotensin type 1 receptor null mice. Neuroscience, 170 (4): 1286-94. [PMID:20727387]
63. Roussy G, Dansereau MA, Doré-Savard L, Belleville K, Beaudet N, Richelson E, Sarret P. (2008) Spinal NTS1 receptors regulate nociceptive signaling in a rat formalin tonic pain model. J Neurochem, 105 (4): 1100-14. [PMID:18182046]
64. Sarrieau A, Javoy-Agid F, Kitabgi P, Dussaillant M, Vial M, Vincent JP, Agid Y, Rostène WH. (1985) Characterization and autoradiographic distribution of neurotensin binding sites in the human brain. Brain Res, 348 (2): 375-80. [PMID:4075096]
65. Schaeffer P, Laplace MC, Savi P, Pflieger AM, Gully D, Herbert JM. (1995) Human umbilical vein endothelial cells express high affinity neurotensin receptors coupled to intracellular calcium release. J Biol Chem, 270 (7): 3409-13. [PMID:7852427]
66. Skrzydelski D, Lhiaubet AM, Lebeau A, Forgez P, Yamada M, Hermans E, Rostene W, Pelaprat D. (2003) Differential involvement of intracellular domains of the rat NTS1 neurotensin receptor in coupling to G proteins: a molecular basis for agonist-directed trafficking of receptor stimulus. Mol Pharmacol, 64 (2): 421-9. [PMID:12869647]
67. Snider RM, Forray C, Pfenning M, Richelson E. (1986) Neurotensin stimulates inositol phospholipid metabolism and calcium mobilization in murine neuroblastoma clone N1E-115. J Neurochem, 47 (4): 1214-8. [PMID:3018165]
68. Tanaka K, Masu M, Nakanishi S. (1990) Structure and functional expression of the cloned rat neurotensin receptor. Neuron, 4 (6): 847-54. [PMID:1694443]
69. Tirado-Santiago G, Lázaro-Muñoz G, Rodríguez-González V, Maldonado-Vlaar CS. (2006) Microinfusions of neurotensin antagonist SR 48692 within the nucleus accumbens core impair spatial learning in rats. Behav Neurosci, 120 (5): 1093-102. [PMID:17014260]
70. Vita N, Laurent P, Lefort S, Chalon P, Dumont X, Kaghad M, Gully D, Le Fur G, Ferrara P, Caput D. (1993) Cloning and expression of a complementary DNA encoding a high affinity human neurotensin receptor. FEBS Lett, 317 (1-2): 139-42. [PMID:8381365]
71. Watson MA, Yamada M, Yamada M, Cusack B, Veverka K, Bolden-Watson C, Richelson E. (1992) The rat neurotensin receptor expressed in Chinese hamster ovary cells mediates the release of inositol phosphates. J Neurochem, 59 (5): 1967-70. [PMID:1328536]
72. White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG et al.. (2012) Structure of the agonist-bound neurotensin receptor. Nature, 490 (7421): 508-13. [PMID:23051748]
73. Yamada M, Yamada M, Watson MA, Richelson E. (1993) Neurotensin stimulates cyclic AMP formation in CHO-rNTR-10 cells expressing the cloned rat neurotensin receptor. Eur J Pharmacol, 244 (1): 99-101. [PMID:8380559]
74. Zhao Y, Wei ZZ, Lee JH, Gu X, Sun J, Dix TA, Wei L, Yu SP. (2020) Pharmacological hypothermia induced neurovascular protection after severe stroke of transient middle cerebral artery occlusion in mice. Exp Neurol, 325: 113133. [PMID:31770520]